Past Issues
Chronic Inflammation and Its Role in Colorectal Cancer Development
Mohsen Mohammadi1, Jalil Mehrzad2*, Nourouz Delirezh1, Abbas Abdollahi3
1Department of Microbiology, Faculty of Veterinary Medicine, Urmia University, Iran
2Department of Microbiology and Immunology, Faculty of Veterinary Medicine, University of Tehran, Iran
3Department of Surgery, Faculty of Medicine, Mashhad University of Medical Sciences, Iran
*Corresponding author: Jalil Mehrzad, Professor of Immunology, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran. Tel: 0021-61117053, Email: [email protected]
Received: May 16, 2022
Published: July 18, 2022
Citation: Mohammadi M, et al. (2022). Chronic Inflammation and Its Role in Colorectal Cancer Development. Oncogen. 5(1):24.
Copyright: Mohammadi M, et al. © (2022).
ABSTRACT
Colorectal cancer (CRC) is the third common cancer and fourth cancer related mortality, worldwide. Chronic Inflammation is one of the underlying mechanisms leads to this cancer. The final aim of this research was to assess the expression level of pattern recognition receptors (PRRs)-related genes TLR2, TLR4, NLRP3, NF-kβ, MBL1, GAL1 and oxidative agent NOS2 in colon tissue and blood monocytes of CRC patients to find the relative role of chronic inflammation in the initiation of the CRC development. By this approach, the expression level of mentioned genes was assessed in colon tissues and monocytes of 24 cases (12 CRC and 12 healthy control persons) with 65±11 years old. In CRC patients’ monocytes, the expression level of TLR2, TLR4, GAL1 and MBL1 genes were significantly less than those of healthy controls (p<0.05). The expression level of NOS2 and NF-kβ genes in CRC patients’ monocytes was significantly higher than those of in healthy controls (p<0.05). In cancerous colon tissues, the expression level of TLR2, TLR4 and NLRP3 was significantly higher and in the case of NOS2 and NF-kβ the difference was not significant. But, the expression level of MBL1 was significantly lower than normal tissues (p<0.05) .In conclusion, since CRC development is normally accompanied by chronic inflammation, the observed significant differences at the expression level of some key inflammatory genes in monocytes and cancerous colon tissues of CRC patients, is highly related to chronic inflammatory reactions in tumor microenvironment.
Keywords: Toll like receptors 2, 4, NLR family pyrin domain containing 3 (NLRP3), Nitric oxide synthase2 (NOS2), Mannose binding lectin1 (MBL1), Galectin1 (GAL1), Nuclear factor kappa binding, beta actin (ACTB), Colorectal cancer (CRC)
INTRODUCTION
Colorectal cancer (CRC) is the third common cancer and forth cancer mortality causative agent, all over the world [1]. Its incidence is increasing in developing countries [2]. On the basis of global cancer statistics, it is the third and second cancer in men and women respectively. Accordingly, 1.2 million new cases and 608700 mortality cases occurred in 2008 [3]. The CRC incidence rate is age related; thus, the highest incidence has been reported in ages 75 and up [4]. Indeed, CRC has been one of the commonest cancers in Iran in year 2008, and also, as GLOBOCAN statistics, it has been in year 2012, the third and fourth common cancer in Iranian men and women respectively [5,6].
Although, CRC within all kinds of cancers, has the most relationship to chronic inflammation, but, the exact relationship of CRC with chronic inflammation remained to be determined. However, in this cancer, the immune dysregulation by inflammatory cells and molecules has been shown [7]. As estimated, approximately 40% of cases, develop to fatal metastasis, of which 80% tend to liver and 20% to other viscera and organs [8,9]. On the basis of broad investigations, the involvement of pattern recognition receptors (PRRs), especially TLRs, has important roles to cancer initiation and development. In this regard, the damage associated molecular patterns (DAMPS) released from damaged epithelial tissues during an infection in colon, have important role in PRRs’ continuous signaling stimulation followed by the creation of chronic and permanent inflammation [10]. Generally, PRRs trigger primary immune responses against infection and tissue damage to save the host from microbial invasion. In this context, it has been shown the significant relationship between TLR4 expression by tumor cells and high risk of tumor recurrence in CRC patients [11]. The recognition of pathogen associated molecular patterns (PAMPs) by PRRs activates several cell signaling that in turn affects the host defense, inflammation development, cell death, amplification and tissue repair. Furthermore, this signaling can activate the dendritic cells to trigger and amplify the anti-tumor immunity. The damage or transformation creates intra epithelial cell induction of PRRs ligands. In this regard, it is possible that, the targeted manipulation of PRRs signaling seems to be a fortune to develop new diagnostic and therapeutic methods for many gastrointestinal malignancies [12]. Therefore, these receptors have central roles in many inflammatory diseases and gastrointestinal tumorigenesis. In response to any tissue damage, The PRR signaling starts the tissue repair process. Recent studies have suggested that because of the role of cancer induction as well as anti-tumor immunity progression for receptors like TLR4, the PRR signaling dysregulation followed by inflammatory damages, can be as a pre-request to intestinal neoplastic changes [11,12]. It is possible that, signaling by these receptors leads to amplification and survival of cancer cells by producing Nuclear Factor-kappa β (NF-kβ). NF-κβ is a ubiquitous transcription factor that mediates a cytoplasmic/nuclear signaling pathway and regulates gene expression of various cytokines, cytokine receptors and adhesion molecules involved in inflammatory and immune reactions. Furthermore, there is a correlation between the activation of NF-κB and control of apoptotic pathway, cell proliferation, differentiation, migration, and angiogenesis as well as resistance to chemo/radiotherapies in tumor cells [13]. The NF-κβ signaling pathway plays an important role in the regulation of cell proliferation and cell survival. Constitutive activation of this pathway leads to the constitutive expression of proliferation-associated genes including cyclin D1, cyclin E, and cyclin-dependent kinase (CDK)-2, as well as IL-6 and Myc. Standing with aberrant regulation of NF-κβ is frequently reported in tumor cells. Hence, the inhibition of this cascade may limit cell proliferation [13].
On the basis of clinical studies, the expression of PRRs, is not restricted to immune cells, but their expression increases in many human epithelial tissue cancers. Thus, the PRRs’ signaling is emerged by double roles, the immune system stimulation and tumor growth augmentation, that followed by homeostasis, chronicity and dysregulation, respectively [14].
Generally, the peripheral blood monocytes’ gene expression profile in CRC patients is different from those of healthy persons, and this difference derived from immune systems’ different reactions against tumor cells [15]. These blood cells initiate the inflammatory responses via phagocytosis, inflammatory cytokine secretion, and producing the free oxygen and nitric oxide radicals within innate immune system (see appendix) [16,17]. Also, the regulation of the inflammatory response homeostasis by monocytes, is important; one of these homeostasis is the decreasing or increasing of the exclusive gene expression rate in the inflammatory responses. Therefore, these cells, besides possessing a role in immune response homeostasis, have important roles in tissue repair and combating against tumor cells such as CRC [17].
Here, we hypothesized that the expression rate of key PRRs, such as, TLR2, TLR4, NLRP3, MBL1, GAL1, and inflammatory agents (e.g., NF-kβ and NOS2) genes in colon tissues and peripheral blood monocytes of CRC patients is different from those in healthy persons. By this approach, the aims of this research was to assess the role of chronic inflammation in promoting CRC by estimating the expression rate of the mentioned key PRRs-related genes and NF-Kβ, NOS2 in colon tissues and blood monocytes of stage II CRC patients against healthy persons referred to Ghaem, Omid and Imam Reza hospitals in Mashhad holy city, Iran by CYBR green qPCR method.
PATIENTS AND METHODS
I- Sampling
This research conducted in Iran, University of Tehran, Faculty of Veterinary Medicine, Department Microbiology and Immunology, matching with three Mashhad hospitals, (Ghaem, Omid, and Imam Reza) approved by the Ethical Committee of Mashhad University of medical sciences by ethical code: ID IR.FUBS.REC.1395.42456 with the commitment of the principles of the declaration of Helsinki. All subjects signed their informed consents before participation (January 2017 to December 2019).
We obtained samples from CRC patients underwent surgery (cancerous colon tissues, n=12, 10 cm adjacent to cancerous colon tissues, n=12, venous blood samples, n=12 from those patients) and healthy non-CRC persons (only venous blood samples, n=12) diagnosed by colonoscopy and CT scan (subsequent confirmed by microscopic examination) [18], with signed consent forms and questionnaires, colonoscopy, and CT scan. Before conducting any sampling, colonoscopy, CT scan, and biopsy of site of the CRC were carried out for early detection and confirmation of the type and the stage of CRC by Tumor-Node-Metastasis (TNM) method [18]. Patients who died during study or underwent to chemotherapy or radiotherapy were excluded. All samples taken from stage II CRC as well as the same gender (all from men) and 65±11 years of age [19,20].
II- Monocyte isolation, RNA extraction and cDNA synthesis from samples
Very briefly, The PBMCs were isolated using density gradient centrifugation (eppendorf) on a Histopaque- 1.077 gradient Ficol. Centrifugation was performed at room temperature, 400 g, for 30 min. Then, cells were washed twice in phosphate-buffered saline (PBS) with 2% fetal calf serum (FCS) and 1 mM EDTA. Subsequently, monocytes were isolated by plastic adhesion method [21]. In this way, The PBMCs were washed twice with cold RPMI- 1640 medium, and then, 5×106 PBMCs /ml were plated in 1 NuclonTM Delta surface treated T-75 cell culture flasks. After incubation for 2 hrs at 37⁰C, 5% CO2, and 95% moisture, the non-adherent cells were removed by thorough washing with RPMI-1640 medium and the adherent monocytes on the bottom of the plate, harvested and resuspended into RPMI-1640 enriched with 100U/ml penicillin, 100µg/ml gentamycine, and 0.3mg/ml L-glutamin [21]. RNA extraction was performed using Trizole reagent. Briefly, by adding 1ml Trizole reagent to any cell pellet ingredients, the microtubes were incubated for 5 min at room temp and then 200µl cold chloroform added them on ice for 5 min and then, centrifuged at 12000rpm 4⁰C. Then, isopropanol was added and incubated for 15 min on ice and again centrifuged at 12000rpm 4⁰C. Finally, the 75% ethanol added to pellet and centrifuged for 5 min at 7500rpm and incubated at room temp to form RNA mass. RNA integrity was then, assessed using an RNA 2000C nanodrop. RNA integrity number was more than 1.6. RNA was solubilized in DEPC treated water and maintained until the next use, at -80⁰C. For each sample, 200 ng of total RNA was reversed transcribed into cDNA using Revert Aid FIRST Strand cDNA synthesis [21].
III- qPCR performing
After primer designing for mentioned genes, by Oligo7 software (DBA Oligo,Inc,USA), and achieving their genuinity by NCBI-BLAST [20, 22-24], The gradient PCR was conducted to obtain the annealing temperatures for primers, and then, the qPCR technique was performed (using Quiagen Rotor gene Q series) by 0.25ng cDNA from each sample, using Yekta Tajhiz Azma Co IRAN, CYBR Green mastermix including 400µmol each dNTPs, 5mMol MgCL2, and 0.1U/ µL Taq DNA polymerase enzyme. In each amplification, the specific primer was used by 0.2µMol. The cycling programs, on the basis of annealing temperature, were as: 10 min at 95⁰C (Hold 1), 40 cycles, 10sec at 95⁰C, 20sec at 61⁰C (for TLR2,TLR4, NLRP3, NOS2 and NF-kβ genes) and 58⁰C( for GAL1, MBL1, and ACTB genes), 20sec at 72⁰C, 1min at 95⁰C (Hold 2), for a total of 40 cycles. All samples were run in duplicate and ACTB gene used as internal reference gene and relative quantification of the total gene products in each sample was done by ∆∆Ct method, as Schmittgen et al. [25].
IV- Statistical Analysis
The data were assessed by SAS statistical software (Version 21.C, USA). Meanwhile, the plots was drown by Graphpad Prism (Soft98 Version, IRAN). Regarding to abnormality of distribution, and also low sample numbers, the Mann-Whitney test was applied for comparing gene expression alterations between CRC and healthy persons, and p<0.05 was considered as significant difference between the groups [21].
RESULTS
As representatively demonstrated, a sample of isolated monocytes stained by Giemsa method is shown in Figure 1:
Figure 1. A sample from Giemsa-stained smear of pure monocytes, isolated by the plastic adhesion technique (×1000).
I- The qPCR results about monocytes genes of interest
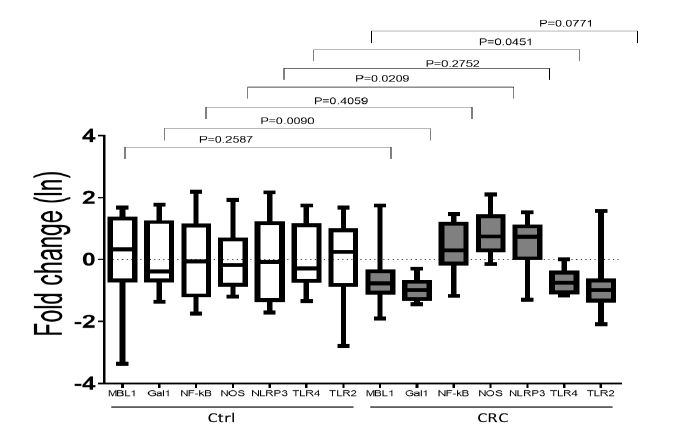
The results of qPCR have shown in figure 2.
Figure 2. The bars of fold change of interested genes expression of isolated monocytes of CRC patients (at right) and healthy controls (at left). As shown by fold change bars, the expression rate of GAL1 and TLR4 genes in CRC comparing to control have significantly decreased (p=0.04 and p=0.009 respectively), NOS2 gene, has significantly increased (p=0.02), but NLRP3 and NF-Kb genes have not significantly decreased (p=0.27 and p=0.40 respectively). On the other hand, the expression rates of MBL1 and TLR2 genes compared to control, were not significant changed (p=0.25 and p=0.77 respectively). The DATA here is on the basis of Naperian logarithm (Ln).
II- The qPCR results about colon tissues
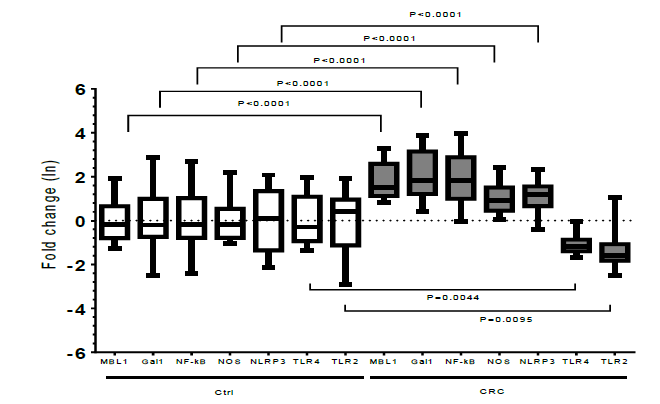
The results have shown in figure 3:
Figure 3. The bars of fold change of interested genes expression of cancerous (at right) compared to adjacent healthy colon tissues (at left). As shown by fold change graphs, the expression of all interested genes in this research significantly differed in cancerous compared to adjacent healthy colon tissues. Thus, the expression rates of TLR2 and TLR4 genes, have significantly decreased, but NLRP3, NF-Kb, NOS2, MBL1, and GAL1 significantly increased (p<0.05). The data here is on the basis of Naperian logarithm (Ln).
DISCUSSION
CRC, is the third common cancer and the forth cancer mortality globally [1]. Since the chronic inflammation due to key PRRs dysregulated hyper or hypo genes’ expression, initiates and promotes some gastrointestinal cancers, such as CRC [7]. In this approach, the estimation of the expression rate of TLR2, TLR4, NLRP3, MBL1, GAL1, NF-Kβ and NOS2 genes in monocytes, cancerous and non-cancerous colon tissues of 12 CRC patients as a beneficial approach to CRC prognosis, early diagnosis, and treatment in its early stages was evaluated in this research.
In monocyte samples of CRC patients, the expression level of GAL1, and TLR4 genes were significantly decreased (p<0.05). This could presumably be related to variable mRNA transcription levels, under some epigenetics conditions, and the mRNAs’ instability in tumor cells because of genome instability. Also, it could be the result of the cell differentiation degree and even the various type of blood monocytes and thus monocytes-derived macrophages; because, in monocyte derived M1 macrophages at tumor microenvironment, the expression level of the mentioned genes can be increased. This phenomenon, maybe, due to monocyte’s exposure to bacterial lipopolysaccharide and peptidoglycan in gut lamina propria and blood [26, 27]. It has been proposed that, the expression level of some genes in blood cells of cancer patients is affected by extracellular vesicles derived from circulatory tumor cells, because of some suppressor cytokine secretion [28]. The expression level of NOS2 gene was significantly increased (p<0.05). The exact role of NOS2 in CRC pathogenesis is still ambiguous and controversial. In previous studies, the expression level of NOS2 gene, was upregulated in tumor samples both at systemic (in leukocytes) and local levels compared to healthy controls, which is significantly associated with the pathological stage [29]. However, other researchers observed higher NOS2 expression in the unaffected mucosa than in colorectal tumor tissues [30]. In our study, also, the expression level of NOS2 gene in cancerous colon tissues was significantly higher than unaffected healthy epithelia (P<0.0001). We suggest that NOS2, has protective effects in this stage. Studies also demonstrated that high expression level of NOS2 and following NO secretion in monocytes-derived M1 macrophages has both cytostatic and cytotoxic roles to CRC cells [31]. In cancerous colon tissues, the significant decreased expression level of TLR2, TLR4 genes was considerable, probably due to deletion mutations in mentioned genes, as reported elsewhere [32]. This issue was relatively due to polymorphisms occurred in these two genes. Also, TLR2 can increase the tendency towards sporadic CRC [18]. Therefore, it seems possible that the decreased expression level of TLR2 and TLR4 in our study, herein, may be due to a kind of negative host defense mechanism in stage II CRC; nevertheless, it can be affected by mutations as well. On the other hand, TLR4 expressed by human cancerous colon cells can induce immunosuppression, as well as, apoptosis resistance, which help tumor to escape from immune system [1]. As such, we suppose the notion that the decreased expression level of TLR4 gene in stage II CRC patients may be beneficial towards decreasing colorectal cancerous cells’ resistance to apoptosis and inhibiting their immune evasion (see appendix).
The significantly increased expression level of both NF-Kβ and NOS2 genes in colorectal cancerous tissues points the notion to the central role for NF-Kβ as a transcription factor for causing inflammation in gut; this is due mainly to the significantly increased expression level of NLRP3 gene, which might possibly be caused by NLRP3 polymorphisms and also, alternative splicing even in its promoter that occurred in this gene and leads to its upregulation [33]; however, it seems possible that this issue can be a compensatory factor against decreased TLR2 and TLR4 genes’ expression level. Also, supposedly the increased level of MBL1 and GAL1 genes in colorectal cancerous tissues, was due to linking feature of these two receptors on the surfaces of cancerous cells to ease metastasis help them invade other tissues. Following complement activation, MBL molecule can link to colon cancerous cells, increasing phagocytosis and dubbing the inflammatory reactions, as explained by Ytting, et al, [34]. Also, The expression level of the GAL1 gene, is increased in cancerous tissues as an apoptosis promoter, as mentioned by Hannah, et al, [35]. Therefore, here it was supposed that the increased level of GAL1 gene is a defense mechanism, by which the tumor cell amplification is inhibited and makes tumor cells more apoptotic.
This research had several limitations, including: 1) the expression level of some inflammatory genes was estimated and other screening factors for CRC such as CEA (carcinoembryonic antigen) was not considered, 2) the expression rate of isolated stages of this cancer was not considered, and 3) involvement of isolated monocytes populations and sex factor was not considered. Therefore, we recommend the study of the mentioned factors and their relationship to the expression levels of the studied genes of interest is worthwhile.
CONCLUSIONS
Considering relationship between the mentioned gene expression and CRC promotion, it seems that, the observed significant up/down-regulation in the expression level of some inflammatory genes in monocytes and cancerous colon tissues of the CRC patients could relate to chronic inflammation and these changes may be benefits to early diagnosis of CRC. But, it needs more investigation.
CONFLICTS OF INTEREST
“The authors declare that there is no conflict of interest regarding the publication of this manuscript”.
ACKNOWLEDGEMENTS
The authors thank Dr. Dr Ali Ghorbani, Dr Mir-Hadi Jazayeri, Dr Hamid Nikkhah for advice on this work and acknowledge the Research Buerue of Faculties of Veterinary Medicine in University of Tehran and Urmia University to support this work.
Appendix
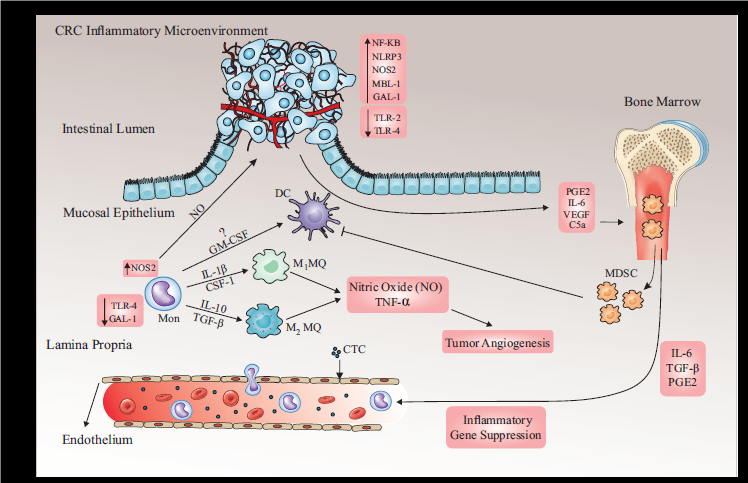
Schematic view of our results-related cellular-molecular involvements in CRC microenvironment. Following tumor cell releasing of CCL2 chemokine, blood monocytes recruit to tumor microenvironment and polarize to dendritic cells (DC), M1 and M2 macrophages depending on the tumor microenvironment. These cells are activated by the specific ligands for TLR2, TLR4 and NLRP3 receptors and inflammatory cytokines such as TNFα, IL-1β, IL-6 and free radicals (eg, H2O2, NO, 2O2-, etc.), leading to tumor cell angiogenesis and DC suppression. Further, the tumor cells and M2 macrophages release suppressor cytokines (eg, TGFβ, PGE2, VEGF and C5a) lead to releasing myeloid derived suppressor cells (MDSCs) from BM followed by releasing IL-6, TGF- β and PGE2 to vascular system as well as tumor microenvironment leading to DC and monocyte suppression in two mentioned milieus respectively; this followed by overexpression of GAL1, TLR4 and NOS2 gene in immune cells specially monocytes. Also, the circulatory tumor cells (CTCs in the picture) in peripheral blood of CRC patients can affect the monocytes’ gene expression profile. On one hand, in cancerous colorectal epithelia overexpression of NF-KB, NLRP3, NOS2, MBL1 and GAL1 along with downregulation of TLR2 and TLR4 in CRC patient happen. However, the cancerous epithelial increasing level of two lectins MBL1 and GAL1 genes can be related to the changes on the tumor cell surface glycosylation template. But, NOS2 gene, by producing nitric oxide synthase consequently produces the nitric oxide, causing angiogenesis which may damages cellular DNA, leading to tumor resistance to apoptosis. Furthermore, the upregulation of NLRP3 gene in colorectal cancerous epithelia may be as a revenging phenomenon against downregulation of TLR2 and TLR4 in epithelia, promoting inflammasome complex, leading to upregulation of NF-KB gene, and thus inflammatory gene transcription and caspase activation, thereby augmenting IL-1β secretion from the colorectal epithelia, possible boosting colorectal tissues’ repair; as such, these double-edged sword cellular and molecular interactions can lead to CRC development.
REFERENCES
- Tang X, Zhu , Wang HBS, Zhu YQ, Wei B (2010). Expression and functional research of TLR4 in human colon carcinoma. Am J Med Sci. 339(4):319-326.
- Center M, Jemal A, Ward E. (2009),International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 18(6):1688-1694.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011). Global cancer statistics. CA Cancer J Clin. 61(2):69-90.
- Boyle P, Leon ME. (2002). Epidemiologyof colorectal cancer. Br Med Bull. 64 (1): 1-25.
- Moghimi-Dehkordi B, Safai A, Zali MR. (2008). Prognostic factors in 1,138 Iranian colorectal cancer patients. Int JColorect Dis. 23(7): 683-688.
- Dolatkhah R, Somi MH, Bonyadi MJ, Asvadi Kermani I, Farassati F, et al., (2015).Colorectal cancer in iran: molecular epidemiology and screening strategies. J Cancer Epidemiol. 643020.
- Rafa H, Benkhelifa S, AitYounes S, Saoula H, Belhadef S, et al. (2017), All-Trans Retinoic Acid Modulates TLR4/NF-kB Signaling Pathway Targeting TNF-alfa and Nitric Oxide Synthase 2 Expression in Colonic Mucosa during Ulcerative Colitis and Colitis Associated Cancer. Mediators of Inflammation. 1-16.
- Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, et al. (2006). Epidemiology and management of liver metastases from colorectal cancer Ann. Surg,. 244(2):254-259.
- Tsikitis VL, Larson DW, Huebner M, Lohse CM, and Thompson PA. (2014). Predictors of recurrence free survival for patients with stage II and III colon cancer. BMC Cancer. 14: 336.
- Gombault A, Baron L, Couillin I. (2013). ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol. (3): 414.
- Eiro NC, Cid JF, Andicoechea S, García-Muñiz A, González JL, et al. (2019). Toll-Like Receptor 4 and Matrix Metalloproteases 11 and 13 as Predictors of Tumor Recurrence and Survival in Stage II Colorectal Cancer. Pathol Oncol Res. 25(4):1589-1597.
- Abreu MF. Fukata M. (2009). Pathogen recognition receptors, cancer and inflammation in the gut. ". Curr Opin Pharmacol. 9(6):680-687.
- Soleimani A, Farzad R, Gordon AF, Mikhai R, Avan A, et al, (2020). Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene. 726:144132.
- Castaño-Rodríguez N, Nadim OK, Mitchell HM. (2014). Pattern-recognition receptors and gastric cancer. Front Immunol. 5: 336.
- Fridman W, Pages F, Saut`es-Fridman C, GalonJ. (2012). The immune contexture in human tumours: impact on clinical outcome. Nature Reviews Cancer. 12(4):298–306.
- Parkin J, Cohen B. (2001). An overview of the immune system. Lancet. 357(9270):1777-1789.
- Parihar A, Eubank TD, Doseff AI. (2010). Monocytes and macrophages regulate immunity through dynamic networks of survival and cell death. J Innate Immun. 2(3):204-215.
- Nihon Y, Terai K, Murano T, Matsumoto T, Okazumi S. (2012), Tissue expression of Toll-like receptors 2 and 4 in sporadic human colorectal cancer. Cancer Immunol Immunother. 61(1):71-77.
- Ghorbani R, Mehrzad J, Dehghani H, Abdollahi A and S Hosseinkhani. (2020). Variation in blood and colorectal epithelia's key Trace elements along with expression of mismatch repair proteins from localized and metastatic colorectal cancer. Biol Trace Elem Res. 194(1):66-75.
- Mehrzad J, Malvandi AM, Alipour M, Hosseinkhani S. (2017). Environmentally relevant level of aflatoxin B1 elicits toxic pro-inflammatory response in murine CNS-derived cells. Toxicology Letters. 279: 96-106.
- Mehrzad J, Bahari A, Bassami MR, Mahmoudid M and Dehghan H. (2018),Immunobiologically relevant level of aflatoxin B1 alters transcription of key functional immune genes, phagocytosis and survival of human dendritic cells. j.imlet. 197:44-52.
- Furrie E, Macfarlane S, Thomson G, Macfarlane G. (2005). Toll-like receptors-2, -3 and -4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology. 115(4):565-574.
- Chien-Chang L, Hsing-Chun K, Feng-Sheng W, Ming-Huey J, Ko-Chao Lee. (2015). Upregulation of TLRs and IL-6 as a Marker in Human Colorectal Cancer. Int. J Mol Sci. 16(1):159-177.
- Malvandi A, Loretelli C, Nasr MB, Zuccotti GV, Fiorina P. (2019). Sitagliptin favorably modulates immune-relevant pathways in human beta cells. Pharmacological Research. 148: 104405.
- Schmittgen TD, Livak KJ. (2008). Analyzing real-time PCR data by the comparative C T method. Nature protocols. 3(6): 1101 -1108.
- Fu-Tong L, Daniel K Hsu, Zuberi RI, Kuwabara I, Chi EY, et al. (1995). Expression and Function of Galectin-3, a beta-Galactoside-Binding Lectin, in Human Monocytes and Macrophages. Amencan Journal of Pathology. 147(4):1016-1028.
- Shimura T, Masahiko Sv, Kenji G, Nakajim T, S Chida, et al. (2016). Association between circulating galectin3 levels and the immunological, inflammatory and nutritional parameters in patients with colorectal cancer. Biomedical reports. 5(2): 203-207.
- Popēna I, Ābols A, Saulīte L, Pleiko K, Zandberga E, et al. (2018), Effect of colorectal cancer-derived extracellular vesicles on the immunophenotype and cytokine secretion profile of monocytes and macrophages. Cell Communication and Signaling. 16(1):1-12.
- Benkhelifa S, Rafa H, Belhadef S, Ait-Kaci H, Medjeber O, et al. (2019). Aberrant up-regulation of iNOS/NO system is correlated with an increased abundance of Foxp3+ cells and reduced effector/memory cell markers expression during colorectal cancer: immunomodulatory effects of cetuximab combined with chemotherapy. Inflammopharmacology. 27(4): 685-700.
- Moochhala S, Chhatwal VJ, Chan ST, Ngoi SS, Chia YW, et al. (1996). Nitric oxide synthase activity and expression in human colorectal cancer. Carcinogenesis. 17(5):1171-1174.
- Ta Lee C, Fang Su P, Lin PC, Tsai HW, Lam CF, et al. (2016). Reappraisal of the significance of inducible nitric oxide synthase in colorectal cancer. J Cell Sci Ther. 7:2.
- El-Abd E, Abou-Shamaa L, Ghoneim H, Fayed W, Al-Wasaby S, et al. (2012). Toll like receptor-4 expression and signaling in patients with colorectal carcinoma. Canadian Journal on Medicine. 3(2):23-29.
- Hoss F, Mueller JL, Ringeling FR, Rodriguez-Alcazar JF, Brinkschulte R, et al. (2019). Alternative splicing regulates stochastic NLRP3 activity. Nature communications. 10(1): 1-13.
- Ytting H, Christensen IJ, Steffensen R, Alsner J, Thiel S, et al . (2011). Mannan‐binding lectin (MBL) and MBL‐associated serine protease 2 (MASP‐2) genotypes in colorectal cancer. Scandinavian journal of immunology. 73(2): 122-127.
- Hannah B, Rhodes JM, Yu LG. (2011). The role ofgalectins in colorectal cancer progression. International Journal of Cancer. 129(1):1-8.
 Abstract
Abstract  PDF
PDF