Past Issues
Targeting Overexpressed Cyclin Dependent Kinase 1 (CDK1) in Human Cancers: Kamalachalcone A Emerged as Potential Inhibitor of CDK1 Kinase Through in Silico Docking Study
Aruna Talatam1, Phani Kumar Reddy2*, Noboru Motohashi3 Anuradha Vanam4, Rao Gollapudi5,*Aruna Talatam1, Phani Kumar Reddy2*, Noboru Motohashi3 Anuradha Vanam4, Rao Gollapudi5,*
1Pulmonary Medicine, NRI Academy of Medical Sciences, Guntur-522503, India
2Internal Medicine, NRI Academy of Medical Sciences, Guntur-522503, India
3Meiji Pharmaceutical University, 2-522-1 Noshio, Kiyose-shi, 204-8588 Tokyo, Japan
4Sri Venkateswara University, Tirupathi-517502, India
5The University of Kansas, Lawrence, Kansas-66045, USA
*Corresponding Authors: Phani Kumar Reddy, Internal Medicine, NRI Academy of Medical Sciences, Guntur-522503. India; Email: [email protected]
Rao Gollapudi, The University of Kansas, Lawrence, Kansas-66045, USA; Email: [email protected]
Received Date: January 5, 2023
Publication Date: February 7, 2023
Citation: Talatam A, et al. (2022). Targeting Overexpressed Cyclin Dependent Kinase 1 (CDK1) in Human Cancers: Kamalachalcone A Emerged as Potential Inhibitor of CDK1 Kinase Through in Silico Docking Study. Oncogen. 6(1):25.
Copyrights: Talatam A, et al. © (2023).
ABSTRACT
Background: CDK1 gene located in human chromosome 10, encodes CDK1 (tyrosine) kinase, overexpressed in multiple cancer cells. Upon phosphorylation of tyrosine kinases by adenosine triphosphate, CDK1 kinase is activated in cell cycle progression. Nonetheless, CDK1 kinase excessively partakes in the development and prognosis of specific types of aggressive cancers. Therefore, CDK1 kinase inhibition therapy is a primary target for the treatment of explicitly expressed CDK1 kinase cancers. At present, dinaciclib is the only FDA approved orphan CDK1 kinase inhibitor drug in the treatment of cancers. In molecular docking study, kamalachalcone A with significant higher binding energy competitively bound to the active receptor site of CDK1 kinase, as comparable to dinaciclib. In this framework, kamalachalcone A could emerge as a potential candidate for restricting the over expression of CDK1 kinase in multiple cancers. Objective: The present study aimed to compare the inhibitory activity of dinaciclib (CDK1 kinase inhibitor drug) with kamalachalcone A, a novel chalcone found in kamala dye. Methods: Dinaciclib and kamalachalcone A were docked on human CDK1 kinase 3D structure in molecular docking using AutoDock 4.2.6, MGL tools and molecular dynamic studies with iMOD server (iMODS). Results: These studies revealed that kamalachalcone A was a competitive CDK1 kinase inhibitor alike dinaciclib. The docking results suggested binding site similarities of dinaciclib and kamalachalcone A. The binding energies of docked dinaciclib and kamalachalcone A complexes with CDK1 kinase were -8.52 kcal/mol and -12.81 kcal/mol, respectively. The docking results were substantiated by molecular simulation studies with iMODS. Conclusion: The in silico docking simulation of dinaciclib and kamalachalcone A suggested that kamalachalcone A is likewise a competitive CDK1 kinase inhibitor.
Keywords: Cancer, CDK1 kinase, cell cycle, dinaciclib, kamalachalcone A, in silico docking, molecular dynamics
INTRODUCTION
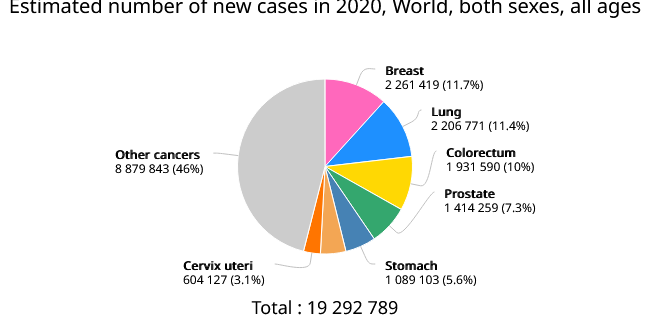
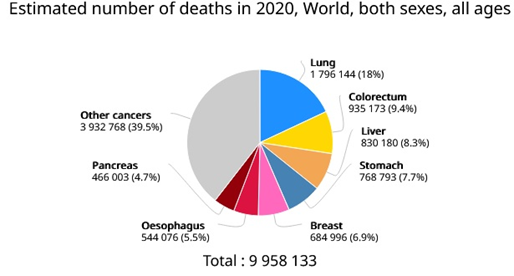
Globally, in this day and age, cancer is reported to be one of the major reasons of deaths and health concern in humans. In 2020, approximately 19.3 million new cancer cases reported worldwide and 10.0 million were cancer related deaths. Cancer incidences and mortalities continue to rise alarmingly in spite of the advancement in diagnostics and therapeutics [1]. Cancers can arise in various body parts resulting in vast range of cancer types and in stage 4, metastasize via angiogenesis to other body parts through blood and lymph systems. In the USA, lung and bronchial cancer mortalities are higher than any other type of cancer [2]. Estimated number of cancer cases and mortalities in 2020 as illustrated in pi chart figure 1 (GLOBCAN 2020).
Figure 1. Estimated number of cancer cases and mortalities in 2020 (GLOBCAN 2020).
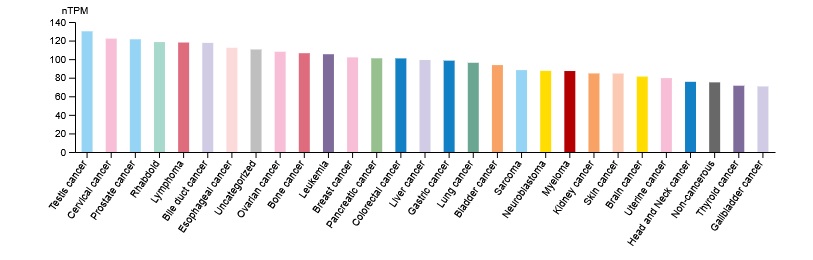
CDK1 gene located in chromosome 10 encodes CDK1 kinase. CDK1 kinase is a member of Ser/Thr protein kinase family. CDK1 kinase is a catalytic subunit of the highly conserved protein kinase complex known as M-phase promoting factor. In cell cycle process, CDK1 kinase activity is regulated by cyclin accumulation and destruction. Furthermore, the phosphorylation and de-phosphorylation of CDK1 kinase play an important regulatory process in cell cycle control [3]. Additionally, CDK1 kinase was especially upregulated by medroxyprogesterone acetate in cancer-associated fibroblasts which resulted in elevated CDK1 kinase expression. Medroxyprogesterone acetate-induced aberrant upregulation of CDK1 kinase might contribute to the enhanced cell proliferation, suggesting a new mechanism of medroxyprogesterone acetate resistance in endometrial cancer [4]. CDK1 kinase expression levels in various cancer cell lines displayed in figure 2.
Figure 2. CDK1 kinase expression levels in various cancer cell lines (The Human Protein Atlas).
The cell cycle process is exceptionally preserved and rigidly regulated which is influenced by checkpoints, cyclin-dependent kinases (CDKs) and cyclins [5, 6]. Any modifications in these functions culminate in the loss of cell cycle control system resulting in uncontrolled proliferation of cells, which eventually induces tumor formation leading to cancers [7-11].
Furthermore, CDK1 kinase overexpression has been found in various cancer types such as breast, colorectal, esophageal adenocarcinoma, gastric, liver, lung, ovarian cancers including oral squamous cell carcinoma [9]. The activity of CDK1 kinase swings in each specific cell cycle, which remains low in G1/S phase and crests at the G2/M phase. Recently, elevated levels of CDK1 kinase expression displayed a prominent activity in stimulating replicative DNA synthesis which resulted in chemo- resistance [12].
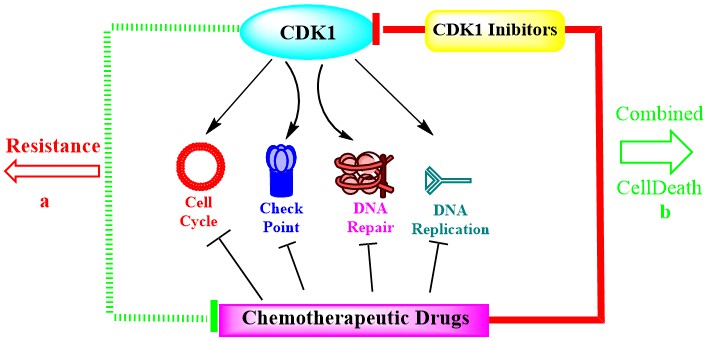
CDK1 kinase is an ideal therapeutic target in drug discovery of adrenocortical carcinoma via regulating epithelial-mesenchymal transition, G2/M phase transition, and apoptosis. [13]. Therefore, targeting cell cycle regulatory proteins is a promising therapeutic strategy in cancer treatments. [3, 14] (Figure 3).
Figure 3: Targets for new drug discovery in cancer treatments.
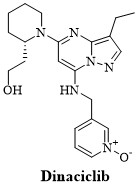
Dinaciclib (figure 4), the orphan drug, is a notably effective CDK1 kinase inhibitor with significant pharmacokinetic properties including safety profile. Likewise, dinaciclib inhibits CDK2, CDK5, and CDK9 kinases and thus reduced cancer progression in broad spectrum of cancer cell lines. [15]. However, dinaciclib treatment displayed certain adverse effects such as leukopenia, alopecia, thrombocytopenia, gastrointestinal symptoms and fatigue [15-19].
Figure 4: Structure of dinaciclib.
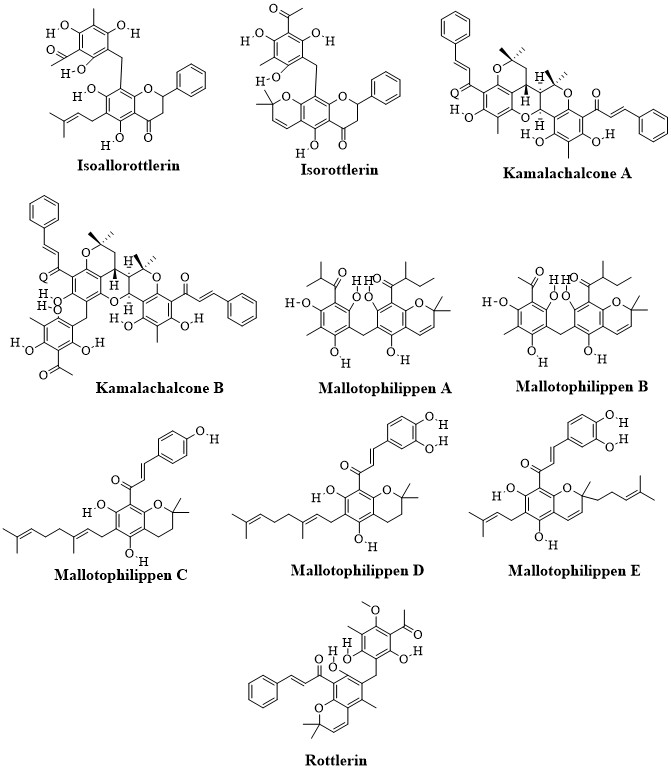
Mallotus philippinensis (Family: Euporbiaceae) comprised a plethora of natural compounds such as phenols, diterpenoids, triterpenoids, steroids, flavonoids, cardenolides, coumarins, and isocoumarins. The chalcones isolated from M. philippinensis such as rottlerin, isorottlerin and mallotophilippinen A-E displayed antimicrobial, antioxidant, antiviral, anti-inflammatory, cytotoxicity, immunoregulatory protein inhibition activity against multiple cancer cells [20-23]. In addition, two novel chalcone derivatives, kamalachalcone A and kamalachalcone B with a distinctive ring system resulted by dimerization between a dimethylchromene ring and a phenoxyl group were isolated from kamala dye (M. philippinensis) [24].
In the present study, chalcone constituents of M. philippinensis, isoallorottlerin, isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E and rottlerin were subjected to molecular docking study to evaluate their inhibitory potency against CDK1 kinase. In addition, the data of these chalcones was compared to dinaciclib along with co-crystallized ligand (FB8301) ((4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine)). Afterwards, the most potent constituent was subjected to molecular dynamic simulation study along with dinaciclib to substantiate the results of molecular docking findings. The structures of isoallorottlerin, isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E and rottlerin are illustrated in figure 5.
Figure 5: Structures of isoallorottlerin, isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E and rottlerin.
MATERIALS AND METHODS
Lipinski’s Rule of Five
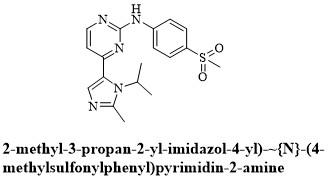
In the beginning stages of drug discovery process, the assessment of drug-likeness of lead molecules helps in reducing experimental costs. The best lead candidate should not violate more than one criterion as defined by the Lipinski's “rule of five”. These rules include molecular weight (no greater than 500 Daltons or g/mol), octanol-water partition coefficient (Logp of 5 or less), hydrogen bond donors (5 or less) and acceptors (10 or less). The drug likeliness tool software (DruLiTo) was used to determine the physicochemical characteristics of isoallorottlerin, isorottlerin, kamalachalcone-A, kamalachalcone-B, mallotophilippen-A, mallotophilippen-B, mallotophilippen-C, mallotophilippen-D, mallotophilippen-E and rottlerin including dinaciclib along with co-crystallized ligand (4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine) (figure 6) with CDK1 kinase (RCSB PDB ID: 6gu7) [25].
Figure 6: Structure of 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine.
Receptor Protein Preparation
The X-ray diffraction crystal structure of CDK1 kinase domain and co-crystalized ligand complex (PDB ID: 6gu7) with 2.75 Å resolution was retrieved from RCSB protein data bank. The protein structure of CDK1 kinase/CDK1 kinase regulatory subunit 2 (Cks) in complex with co-crystallized ligand and co-crystallized water was responsible for elevated resolution and stability of protein. Hence, water molecules, co-crystallized ligand, Cks2 and CDK 1 chains - C, E & G were removed using Discovery Studio software retaining chain A for docking study.
Ligands and Target Preparations
The two dimensional structures of isoallorottlerin, isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E, rottlerin, dinaciclib along with co-crystallized ligand were designed with the help of ChemDraw 21.0 and later, converted to 3D structure using Chem3D 21.0 software.
Docking Between CDK1 Kinase and Ligands
Molecular docking studies were achieved with the help of AutoDock 4.2.6 software so that the Kollman partial charges were allocated to each specific atom of the protein in PDB format. Molecular docking of ligands with CDK1 kinase was conducted to reveal the best sites for the bonding of receptor- co-crystallized ligand and to determine the most stable free energy of receptor-ligand. Three grid boxes with dimensions of X=60 x Y=60 x Z=60, spacing= 0.500Ao with center grid box with x=33.102, y=14.544 and z=10.604 coordinates were used to create grid parameter file (gpf) to cover the active binding site of the receptor molecule followed by the generation of docking parameter file (dpf). These files were used as input files for grid mapping and docking. The PDB files of receptor and ligands were used to generate PDBQT files to execute docking study with AutodDock tools 1.5.7. An independent run was conducted for each ligand and each apparent conformer was docked in a randomized order into the binding pocket of CDK1 kinase. In docking study, genetic algorithm and Lamarckian generic algorithm were assigned for ligand conformational search. The data obtained from AutoDock4 docking log file (dlg) of each docked receptor ligand complex was analyzed [26]. Subsequently, LigPlot+ v.4.5.3 software was used to compute the number of hydrophobic and hydrogen bonds between ligands and CDK1 kinase in the binding site. The type and number of amino acids that exist in the binding site were identified by LigPlot + 4.5.3 software [27].
Molecular Dynamics Simulation Study
The exact internal coordination ameliorates the effectiveness of normal mode analysis while extending its application and preserving the stereochemistry of macromolecule. Vibrational analysis, motion animations including altering trajectories were achieved interactively at divergent resolutions. In this framework, iMODS offers enhanced imagining which clarifies collective motions and better affine-model-based arrow portrayal of domain dynamics. The binding effects of ligands on CDK1 kinase were estimated basing on conformational stability of protein/ligand complex interactions with normal mode analysis (NMA) using iMODS. During this time, several properties such as Deformability, Bfactor, Eigenvalue, Variance, Residue Index and Atom Index of protein/ligand interactions were evaluated [28,29].
RESULTS
Lipinski’s rule of five
The drug likeliness tool software (DruLiTo), used to determine the physicochemical characteristics revealed the molecular weight, Logp, H-donors and H acceptors of isoallorottlerin , isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E, rottlerin and dinaciclib along with co-crystallized ligand with CDK1 kinase. (RCSB PDB ID: 6gu7) Of these compounds kamalachalcone A complied with majority of Lipinski criteria. (Table 1).
Table 1: Lipinski rule of five for ligands.
|
Ligand Name |
Mol. Wt (≤ 500) |
Logp (≤ 5) |
H-Donors (≤ 5) |
H-Acceptors (≤ 10) |
|
Isoallorottlerin |
518.55 |
4.35 |
5 |
8 |
|
Isorottlerin |
516.18 |
3.719 |
4 |
8 |
|
Kamalachalcone A |
656.76 |
8.084 |
3 |
7 |
|
Kamalachalcone B |
836.92 |
8.953 |
6 |
11 |
|
Mallotophilippen A |
498.23 |
4.673 |
5 |
8 |
|
Mallotophilippen B |
470.19 |
4.127 |
5 |
8 |
|
Mallotophilippen C |
476.23 |
6.39 |
3 |
5 |
|
Mallotophilippen D |
492.25 |
6.527 |
4 |
6 |
|
Mallotophilippen E |
490.24 |
6.229 |
4 |
6 |
|
Rottlerin |
528.21 |
5.304 |
3 |
7 |
|
Dinaciclib |
396.23 |
11.643 |
0 |
6 |
|
Ligand (FB8301) |
371.14 |
0.626 |
0 |
7 |
Docking Study
The docked results of ten naturally occurring chalcones: isoallorottlerin, isorottlerin, kamalachalcone A, kamalachalcone B, mallotophilippen A, mallotophilippen B, mallotophilippen C, mallotophilippen D, mallotophilippen E and rottlerin including dinaciclib and ligand with CDK1 kinase listed in table 2.
Table2. Ligands and CDK1 kinase binding energies, inhibitory constants and hydrogen bonds
|
Ligand Name |
Binding Energies (kcal/mol) |
Inhibitor Constant |
Hydrogen Bonds |
|
Isoallorottlerin |
-9.01 |
249.84 nM |
Lys33, Glu81, Leu83 |
|
Isorottlerin |
-9.44 |
121.30 nM |
Lys 33, Gln132 |
|
Kamalachalcone-A |
-12.81 |
405.45 pM |
Leu83, Lys89 |
|
Kamalachalcone-B |
-11.06 |
7.82 nM |
Leu83, Lys89, Gln132, |
|
Mallotophilippen-A |
-8.04 |
1.29 uM |
Leu83 |
|
Mallotophilippen-B |
-8.20 |
980.34 nM |
Lys33, Asp146 |
|
Mallotophilippen-C |
-8.28 |
853.60 nM |
Leu83, Asn133 |
|
Mallotophilippen-D |
-8.56 |
530.08 nM |
Glu8, Lys20, Leu83, Lys89, |
|
Mallotophilippen-E |
-9.10 |
213.72 nM |
Lys33, Asp86, Lys89, Asn133, Asp146 |
|
Rottlerin |
-9.11 |
208.58 nM |
Lys33, Leu83, Asp146 |
|
Dinaciclib |
-8.52 |
3.95 uM |
Leu83, Lys89, Gln132 |
|
Ligand (FB8301) |
-8.14 |
1.08 uM |
Leu83, Asp86 |
mM = Micro-molar; nM = Nano-molar; pM= Pico-molar
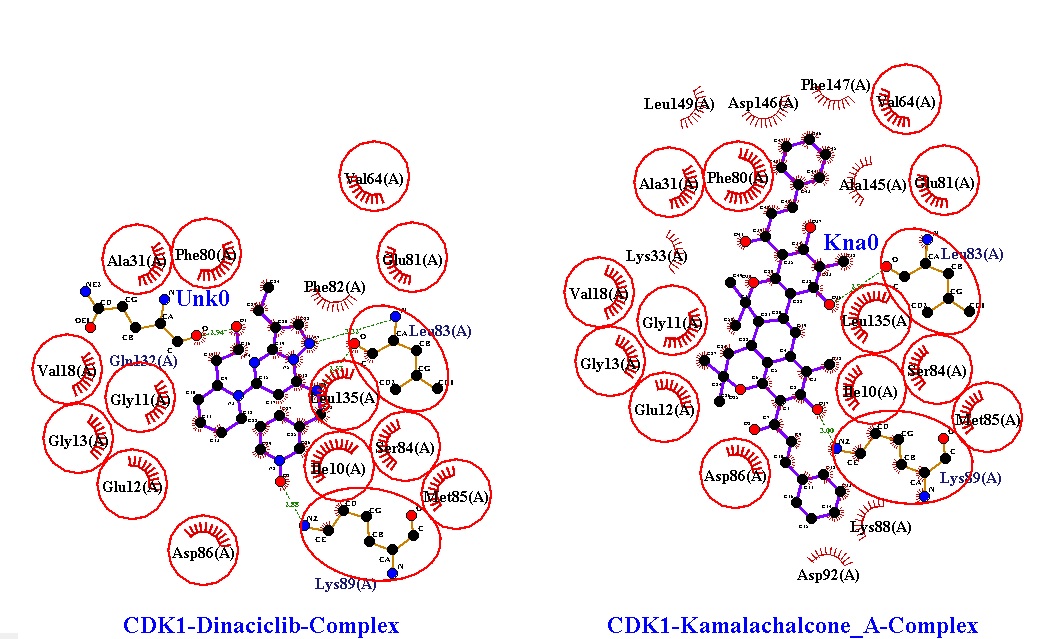
The results from molecular docking of dinaciclib and kamalachalcone A with CDK1 kinase was used as a means to reveal the best sites for the interactions of ligand to receptor to specify the most stable and free energy of ligand/receptor along with dinaciclib. Dinaciclib formed hydrogen bonds at Leu83 (3.01 Ao), Lys89 (2.79 Ao) and Gln132 (2.87 Ao) as well as hydrophobic interactions at Ile10, Gly11, Glu12, Gly13, Val18, Ala31, Val64, The80, Glu81, Ser84, Met85, Asp86, Leu135 amino acid residues of CDK1 kinase. Kamalachalcone A formed hydrogen bonds at Leu83 (2.97 Ao) and Lys89 (3.00 Ao) as well as hydrophobic interactions at Ile10, Gly11, Glu12, Gly13, Val18, Ala31, Lys33, Val64, The80, Glu81, Ser84, Met85, Asp86, Lys88, Asp92, Leu135, Ala145, Asp146, Phe147, Leu149 amino acid residues of CDK1 kinase. Nonetheless, Interestingly, kamalachalcone A formed additional hydrophobic interactions at Lys33,Ala145, Lys88 Asp92, Asp146, Phe147 , Leu149 with CDK1 kinase (figure 7).
Figure 7: Hydrogen bonds and hydrophobic interactions of dinaciclib and kamalachalcone-A with CDK1 kinase.
Molecular Dynamic Simulation Study
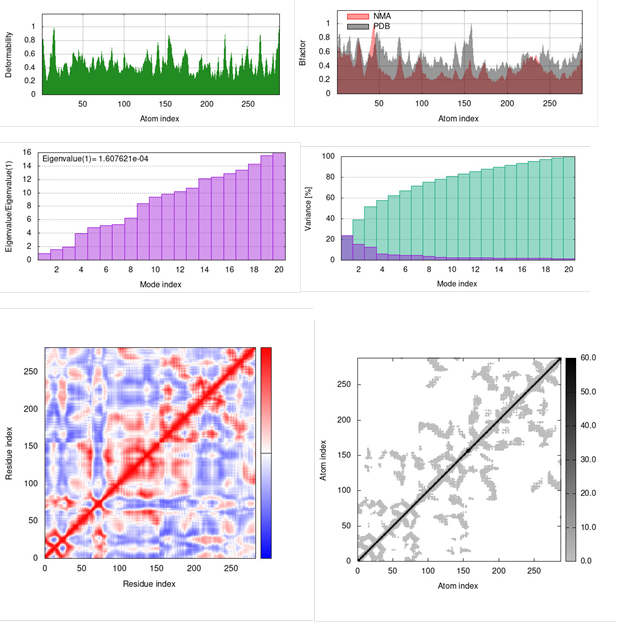
Internal coordinate normal mode analysis server (IMODS) data suggested that a higher covariance matrix indicated a better correlation of protein ligand complex residues. While iMODS is designed to evaluate macromolecules, it is also used to study the stability of macromolecules/small ligand complexes [30, 31]. The iMODS revealed the internal coordinates analysis basing on the protein/ligand structural interactions. In addition, iMODS calculate the B-factor and structural deformity and compute the Eigen value. A good correlation between residues indicated in red color, while the lack of correlation indicated in white color and anti-correlation represented in blue color. The analysis for comparing dinaciclib and kamalachalcone A complexed with CDK1 kinase established that kamalachalcone A displayed equally good correlation comparable to dinaciclib. The energies required for CDK1 kinase complexes with dinaciclib and kamalachalcone A are almost identical. The binding interactions of dinaciclib and kamalachalcone A induced almost similar changes in CDK1 kinase receptor. These statistics of molecular dynamics simulation generated by external iMODS suggest that kamalachalcone A could be a better alternative for dinaciclib. Kamala dye is less toxic bestowed with lesser adverse effects and is more potent in its mechanism of action against human cancer cell lines [32]. The dynamic simulation results of dinaciclib /CDK1 kinase complex are illustrated in figure 8.
Figure 8: Dinaciclib and CDK1 kinase complex: Deformability, Bfactor, Eigenvalue , Variance, Residue Index and atom index.
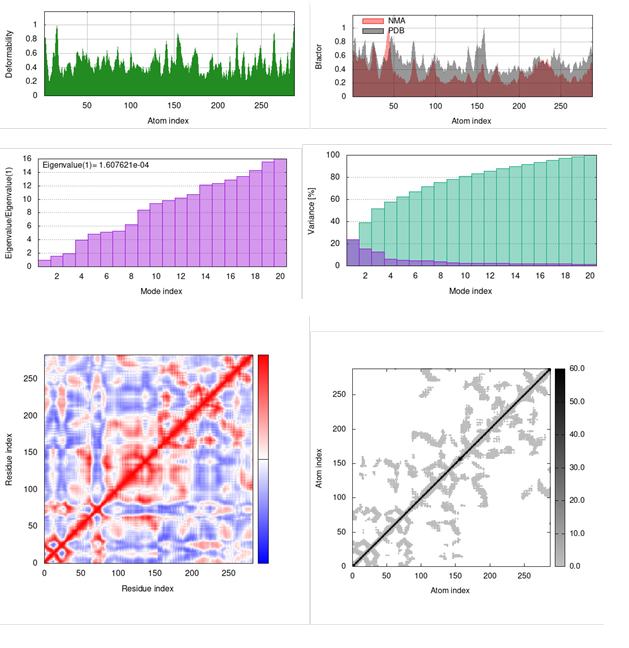
The dynamic simulation results of Kamalachalcone A /CDK1 kinase complex are illustrated in figure 9.
Figure 9: kamalachalcone A and CDK1 kinase complex: Deformability, Bfactor, Eigenvalue , Variance, Residue Index and Atom Index.
DISCUSSION
Dinaciclib is a novel CDK1 kinase inhibitor as well as CDK2, CDK5 and CDK9 inhibitor with prominent influence on multiple cancers in vitro and in vivo. Dinaciclib exhibited promising single-agent activity in myelomas and hence its application in breast cancer treatment was investigated [33]. Dinaciclib attenuated the expression of CDK1 kinase induced anaphase catastrophe in lung cancer [34]. Dinaciclib alone actively induced cell proliferation inhibition, cell cycle arrest and ameliorated the apoptosis along with variations in the expressions of cell cycle and cell apoptosis-related proteins including CDK1 kinase, cyclin D3, cleaved PARP and caspsase-3 in lymphoma Raji cells. [35]. Rottlerin emerged to have great importance for being used in chemotherapy. Thus, rottlerin could be a potent molecule to treat cancers as it would influence cell machineries involved in apoptosis, survival, and autophagy. Furthermore, rottlerin displayed JAK2 inhibitory activity in docking study [21,36].
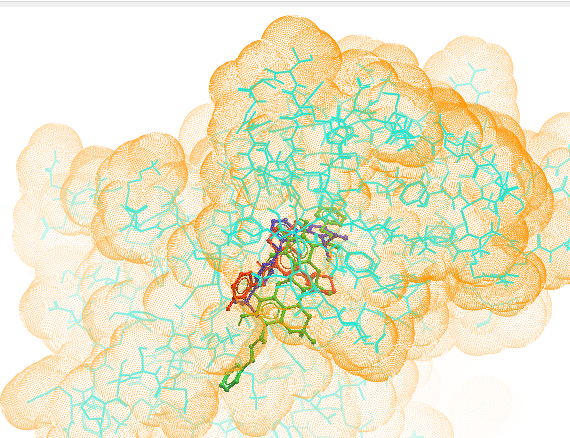
Therefore, the objective of this study is to evaluate potential lead molecules that exhibit significant inhibitory activity of over expressed CDK1 kinase to treat multiple cancers. The active binding site of CDK1 kinase was identified through the observation of ligand binding interactions with CDK1 kinase in CDK1 kinase/ligand complex X-ray crystallographic structure retrieved from Protein Data Bank. In this docking study, the hit molecule, kamalachalcone A was meticulously assessed for its binding ability and behavior in CDK1 kinase active site. The subsequent best pose complexes of dinaciclib and kamalachalcone A from docking was put through molecular dynamics simulation analysis (iMODS) to further evaluate its efficacy in cancer therapies. The data suggested that the hit molecule, kamalachalcone A performed almost in accordance with dinaciclib. Besides, ring generator (Rg) and potential energies advocated compact and consistent protein-hit complexes throughout the simulation process. The residues Leu 83 and Lys 89 located in CDK1 kinase cavity constructed polar interactions with kamalachalcone A surrounded by hydrophobic contacts (figure 7). Interestingly, the excessive hydrophobic interactions of kamalachalcone A with amino acids residues of CDK1 kinase suggest higher binding energy -12.81 kcal/mol while it was -8.52 kcal/mol of dinaciclib. Probably, these excessive hydrophobic interactions of kamalachalcone A contributed to a better alignment of kamalachalcone A in the active cite of CDK1 kinase. The binding patterns and alignments of kamalachalcone A and dinaciclib to CDK1 kinase were almost similar suggesting that kamalachalcone A could emerge as an efficient inhibitor of CDK1 kinase. The alignment of kamalachalcone A, dinaciclib and ligand in the active binding site of CDK1 kinase illustrated in figure 10.
Figure 10: Alignment of kamalachalcone A (green), dinaciclib (red) and ligand (royal blue) in the active binding site of CDK1. (Molsoft browser).
Since kamalachalcone A displayed almost similar interactions with CDK1 kinase receptor comparable to dinaciclib, the graphics of Deformability, Bfactor, Eigenvalue, Variance, Residue Index and Atom Index (figure 8 & figure 9) are almost identical in iMODS simulations study. The statistics of molecular dynamics simulation generated by external iMODS for Deformability, Bfactor, Eigenvalue, Variance, Residue Index and Atom Index effects of kamalachalcone A and dinaciclib complexes with CDK1 kinase are almost similar. These findings suggest that kamalachalcone A is an effective CDK1 kinase inhibitor comparable to dinaciclib. The main chain deformability is a degree of ability of CDK1 kinase to deform at each of its residues. The influence of kamalachalcone A and dinaciclib on the deformability of CDK1 kinase is almost identical. Eigenvalues denotes motion stiffness and is related to the energy required to deform the CDK1 kinase structure. The lower eigenvalues suggest effortless deformation. The eigenvalues of kamalachalcone A and dinaciclib are identical (1.607621e-04) which suggests that kamalachalcone A could be equally potent as dinaciclib. Variance connected to each normal mode is conversely associated to eigenvalue. The variance factor linked to the modes designates their relative contribution to equilibrium motions. Residue index indicates CDK1 kinase correlated, uncorrelated or anti-correlated movements. Elastic network outlines the connection of atoms by springs known as linking matrix of CDK1 kinase. The graphics generated by iMODS for kamalachalcone A and dinaciclib docked with CDK1 kinase displayed nearly identical behavior. Furthermore, the computed inhibitory constant of kamalachalcone A is 405.45 picomolar (pM) compared to dinaciclib inhibitory constant of 3.95 micromolar (mM). This observation suggests that further investigations of CDK1 kinase inhibitory effect of kamalachalcone A in in vitro and in vivo conditions are required to establish kamalachalcone A efficacy in inhibiting over-expressed CDK1 kinase in human cancer cells.
Conspicuously, Lipinski’s rule of five does not apply to the substrates of biological transporters or natural products. Certain high profile drugs such as lipitor and singulair fail more than one of the Lipinski's rules. However, the mean molecular properties of new pharmacological compounds are still credited Lipinski compliant in spite of the fact that their property distributions are distant from historical standards Hence, widely used rules for drug for new drug discovery do not have to comply with Lipinski’s rule of five [37]. In this context, even though kamalachalcone A violate Lipinski’s rule with a higher molecular weight of 656.76 and logp value 8.084 should be considered for the evaluation of its effectiveness as CDK1 kinase inhibitor.
CONCLUSION
Cancer reigns to be the second most common cause of death in the United States after heart disease. Computational approaches have been utilized for discovery and development of new drugs as multi-targeted inhibitors in diseases caused by overexpressed proteins. The present study focused on targeting aberrant levels of CDK1 kinase which is responsible for uncontrolled tumor cell growth. Molecular docking of CDK1 kinase with kamalachalcone A revealed that kamalachalcone A formed complex with CDK1 kinase with binding energy of -12.81 kcal/mol while it was -8.52 kcal/mol for dinaciclib. Thus, kamalachalcone A exhibited significantly higher binding affinity towards the active binding site of CDK1 kinase comparable to FDA approved orphan drug, dinaciclib. Therefore, kamalachalcone A could be a promising lead candidate helpful in the treatment of CDK1 kinase overexpressed cancer for the prevention of cell cycle progression. Consequently, in vitro and in vivo evaluation of kamalachalcone A as a novel CDK1 kinase inhibitor is recommended.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest regarding the publication of this manuscript.
ACKNOWLEDGMENTS
R.G. is grateful to Late Professor Lester A. Mitscher, Late Professor Delbert M Shankel and Late Professor Andrew M Torres in The University of Kansas (KU), Lawrence, Kansas, USA for introducing to computational docking studies at the earlier stages of drug discovery.
REFERENCES
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 71(3):209-249.
- Ridge CA, McErlean AM, Ginsberg MS. (2013). Epidemiology of lung cancer. Semin Intervent Radiol. 30(2):93-98.
- Wiinen R, Pecoraro C, Carbone D, Fiuji H, Avan A, Peters GJ, et al. (2021). Cyclin Dependent Kinase-1 (CDK-1) Inhibition as a novel therapeutic strategy against pancreatic ductal adenocarcinoma (PDAC). Cancers (Basel). 13(17):4389.
- Omar IS, Mat Adenan NA, Godoy A, Teo IH, Gunasagran Y, Chung I. (2022). Aberrant upregulation of CDK1 contributes to medroxyprogesterone acetate (MPA) resistance in cancer-associated fibroblasts of the endometrium. Biochem Biophys Res Commun. 628:133-140.
- Mehraj U, Sofi S, Alshehri B, Mir MA. (2022). Expression pattern andprognostic significance of CDKs in breast cancer: an integrated bioinformatic study. Cancer Biomark. 34:1-15.
- Panagopoulos A, Altmeyer M. (2021). The hammer and the dance of cell cycle control. Trends Biochem Sci. 46(4):301-314.
- Dang F, Nie L, Wei W. (2021). Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 28(2):427-438.
- Mehraj U, Dar AH, Wani NA, Mir MA. (2021 ) Tumor microenvironment promotes breast cancer chemoresistance. Cancer Chemother Pharmacol. 87(2):147-158.
- Li M, He F, Zhang Z, Xiang Z, Hu D. (2020). CDK1 serves as a potential prognostic biomarker and target for lung cancer. J Int Med Res. 48 (2) 300060519897508. doi: 10.1177/0300060519897508
- Sofi S, Mehraj U, Qayoom H, Aisha S, Asdaq SMB, Almilaibary A, et al. (2022) Cyclin-dependent kinases in breast cancer: expression pattern and therapeutic implications. Med Oncol. 39(6):1–16.
- Malumbres M, Barbacid M. (2009). Cell cycle CDKs and cancer: a changing paradigm. Nat Rev Cancer 9(3):153-166.
- Liao H, Ji F, Ying S. (2017). CDK1: beyond cell cycle regulation. Aging. 9(12):2465.
- Ren L, Yang Y, Li W, Zheng X, Liu J, Li S, et al. (2022). CDK1 serves as a therapeutic target of adrenocortical carcinoma via regulating epithelial-mesenchymal transition, G2/M phase transition, and PANoptosis. J Transl Med. 20(1):444.
- Otto T, Sicinski P. (2017). Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 17(2):93-115.
- Lin SF, Lin JD, Hsueh C, Chou TC, Wong RJ. (2017). A cyclin-dependent kinase inhibitor, dinaciclib in preclinical treatment models of thyroid cancer. PLoS One. 12(2):e0172315.
- Feldmann G, Mishra A, Bisht S, Karikari C, Garrido-Laguna I, Rasheed Z, et al. (2011). Cyclin-dependent kinase inhibitor Dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol Ther. 12(7):598-609.
- Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R, et al. (2015). Mayo Phase 2 Consortium. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 125(3):443-448.
- Chen XX, Xie FF, Zhu XJ, Lin F, Pan SS, Gong LH, et al. (2015). Cyclin-dependent kinase inhibitor dinaciclib potently synergizes with cisplatin in preclinical models of ovarian cancer. Oncotarget. 6:14926-14939.
- Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. (2010). Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 9 (8):2344-2353.
- Ma J, Hou Y, Xia J, Zhu X, Wang ZP. (2018). Tumor suppressive role of rottlerin in cancer therapy. Am J Transl Res. 10(11):3345-3356.
- Gangwar M, Goel RK, Nath G. (2014). Mallotus philippinensis Muell. Arg (Euphorbiaceae): ethnopharmacology and phytochemistry review. Biomed Res Int. 2014:213973.
- Lounasmaa M, Widén CJ, Tuuf CM, Huhtikangas A. (1975). On the phloroglucinol derivatives of Mallotus philippinensis. Planta Med. 28(1):16-31.
- Nguyen TM, Chau VM, Phan VK, Nguyen HK, Nguyen XC, Nguyen HD, Nguyen PT, Nguyen HN, La DM, Ninh KB. (2010). Study on chemical constituents of the leaves of Mallotus philippensis. Tap Chi Hoa Hoc. 48:352-357.
- Tanaka T, Ito TT, Iinuma M, Takahashi Y, Naganawa H. (1988). Dimeric chalcone derivatives from Mallotus philippinensis. Phytochemistry. 48(8):1423-1427.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 46:3-26.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Computational Chem. (14):1639-1662.
- Roman A. Laskowski RA, Swindells MB. (2011). LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J Chem Inf Model. 51(10):2778-2786.
- Lopez-Blanco JR; Garzon JI, Chacon P. (2011). iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 27:2843-2850.
- Kovacs JA; Chaco P, Abagyan R. (2004). Predictions of Protein Flexibility: First-Order Measures. Proteins. 56:661-668.
- Zabihollahi R, Namazi R, Aghasadeghi MR, Esfahani AF, Sadat SM, Modarressi MH. (2012). The in vitro anti-viral potential of Setarud (IMOD™), a commercial herbal medicine with protective activity against acquired immune deficiency syndrome in clinical trials. Indian J Pharmacol. 44(4):448-553.
- Sengupta S, Bhowmik R, Acharjee S, Sen S. (2021). In-Silico Modelling of 1- 3- [3-(Substituted Phenyl) Prop-2-Enoyl) Phenyl Thiourea Against Anti-Inflammatory Drug Targets. Biosci Biotech Res Asia. 18(2):413-421.
- Sharma V. (2011). A polyphenolic compound rottlerin demonstrates significant in vitro cytotoxicity against human cancer cell lines: isolation and characterization from the fruits of Mallotus philippinensis . J. Plant Biochem. Biotechnol. 20:190-195.
- Criscitiello C, Viale G, Esposito A, Curigliano G. (2014). Dinaciclib for the treatment of breast cancer. Expert Opin Investig Drugs. 23(9):1305-1312.
- Danilov AV, Hu S, Orr B, Godek K, Mustachio LM, Sekula D, Liu X, Kawakami M, Johnson FM, Compton DA, Freemantle SJ, Dmitrovsky E. (2016). Dinaciclib Induces Anaphase Catastrophe in Lung Cancer Cells via Inhibition of Cyclin-Dependent Kinases 1 and 2. Mol Cancer Ther. 15(11):2758-2766.
- Zhao H, Li S, Wang G, Zhao W, Zhang D, Wang F, Li W, Sun L. (2019). Study of the mechanism by which dinaciclib induces apoptosis and cell cycle arrest of lymphoma Raji cells through a CDK1-involved pathway. Cancer Med. (9):4348-4358.
- Vadapalli, J, Vanam A, Motohashi N, Gollapudi R. (2018). Integrated in Silico Docking and MOMA Simulation Methods Reveal Rottlerin as a Potent Janus kinase 2 (JAK2) Inhibitor. Biomed J Sci Tech Res. 11(1):8216–8225.
- Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. (2012). Quantifying the chemical beauty of drugs. Nat Chem. 4(2):90-98.
 Abstract
Abstract  PDF
PDF