Past Issues
CRISPR/Cas9-based Editing of CDK4, p107, and TGFβ1 in Human Breast and Lung Cancer Cells
Hussein Sabit1*, Doha Alaa2, Nourhan Jamal2, Sodfa Tarek2, Magda Fouda2, Bassant Maher2, Abdel-Bary Prince3, Shimaa E Abdel-Ghany2, Emre Cevik1, Huseyin Tombuoglu1, Osama A M Said2, Abdulrahman Sabry4, Afnan F Al-Muhanna5, Mokhtar El-Zawahri2,6
1Department of Genetics, Institute for Research and Medical Consultations, Imam Abdulrahman Bin Faisal University, Saudi Arabia 2College of Biotechnology, Misr University for Science and Technology, Egypt 3College of Vet. Medicine, Cairo University, Egypt 4GIS Research Center, Egypt 5 Breast Imaging Division, KFHU, Imam Abdulrahman Bin Faisal University, Saudi Arabia 6Center for Research and Development, Misr University for Science and Technology, Egypt
Corresponding author: Hussein Sabit, Department of Genetics, Institute for Research and Medical Consultations, Imam Abdulrahman Bin Faisal University, Saudi Arabia.
Received: May 16, 2019 Published: June 26, 2019
ABSTRACT
The CRISPR/Cas9 system is considered one of the most controversial yet powerful genome-editing tool with high potentiality and wide-angled array of biomedical applications. Cancer is considered one of the serious health-threat diseases that could be controlled, and even, cured by CRISPR/Cas9. In the present study, three cancer-related genes (CDK4, p107, and TGFβ1) were targeted in breast cancer cells (MCF-7) and lung cancer cells (A549) with CRISPR/Cas9 cassettes. Cells were transfected with these crRNAs to knockout CDK and TGFβ1, and to activate p107 in a dose-optimized manner. Viable cell count was measured using Trypan blue assay. Semi-quantitative PCR was also performed to detect the knockout and activation of the genes under study. Two-dimensional polyacrylamide gel electrophoresis was carried out to ensure the up/downregulation of the studied genes in both MCF-7 and A549. Transfecting MCF-7 and A549 has resulted in a significant reduction in the cell count (p>0.005). Cell viability was also measured using MTT assay, and the results showed a significant decrease in the overall viability of cells after being challenged with CRISPR/Cas9. Semi-quantitative PCR was performed against specific primers to ensure the elimination/activation of the target genes. Data indicated a downregulation of CDK4 and TGFβ1 and upregulation of p107. Two-dimensional protein gel electrophoreses was also conducted to indicate the level of up/down regulation of the specific proteins encoded by these genes. Data indicated a partial presence of CDK4 and p107 and a complete absence of TGFβ1 protein. This study might indicate the efficiency of editing tumor-related genes within the genome of malignant cells. However, further confirmative studied are needed to validate the use of CRISPR Knockout (KO)/Activation (AC) in controlling cancer in vitro.
Keywords: CRISPR; Knockout; CDK4; p107; TGFβ1; Lung; Breast; Cancer
INTRODUCTION
Cancer is a leading cause of death worldwide, accounting for 8.8 million deaths in 2015. The most common causes of cancer death are cancers of breast (571,000 deaths) and lung (1.69 million deaths) [1]. Cancer is caused by a series of alterations in genome and epigenome mostly resulting in activation of oncogenes or inactivation of cancer suppressor genes [2, 3]. This disease is a multistep process in which not one, but several mutations are required to cause functional abnormality that leads to tumorigenesis [4].
Breast cancer, in this context, is emerging by a multistep process, which can be broadly equated to transformation of normal cells via the steps of hyperplasia, premalignant change and in situ carcinoma [5]. Several genes are involved in the tumorigenesis and progression of breast cancer, but functional validation of candidate cancer genes remains unsolved [6].
Lung cancer, which has a low survival rate, is a leading cause of cancer-associated mortality worldwide. Smoking and air pollution are the major causes of lung cancer; however, numerous studies have demonstrated that genetic factors also contribute to the development of lung cancer [7, 8]. The overall survival rate of lung cancer patients remains poor despite the availability of standard treatments [9].
Clustered regularly interspaced short palindromic repeats (CRISPR) is a powerful genome editing technique [10]. Cancer characterization and modeling have benefitted greatly from the genome editing capabilities of CRISPR/Cas9 [11]. By rapidly introducing genetic modifications in cell lines, organs and animals, CRISPR/Cas9 system extends the gene editing into whole genome screening, both in loss-of-function and gain-of-function manners [12, 13]. This technology, developed based on the discovery that DNA double strand breaks (DSBs), could stimulate endogenous DNA repair machinery, mainly through homology-directed repair (HDR) and non-homologous end-joining (NHEJ) [14]. Recent CRISPR screens detected thousands of essential genes required for cellular survival and key cellular processes; however, discovering novel lineage-specific genetic dependencies from the many hits remains a challenge [15].
Cellular proliferation is controlled by several cell-cycle checkpoint proteins. In malignancy, the genes encoding these proteins are often disrupted and cause unrestrained cancer growth. The proteins are over-expressed in numerous malignancies; thus, they are potential targets for anti-cancer therapies [16]. One of these proteins becomes the master regulators, cyclin-dependent kinases (CDKs), which serves as the actual driving forces behind the progression of cell cycle in eukaryotic cells [17]. CDK/cyclin complexes regulate each phase of the cell cycle and the breakdown of this regulation in any phase results in uncontrolled growth and thus, tumor formation. Most, if not all of the cancers show direct or indirect deregulation of these kinases, therefore targeting CDKs is an important aim or factor to develop new anticancer therapeutics [18].
The retinoblastoma (RB) tumor suppressor and its family members, p107 and p130, function by repressing E2F transcription factor activity to attenuate the expression of genes essential for cell cycle progression through modulation of the E2F family of transcription factors [19]. Transforming growth factor-beta1 (TGFβ1) is a cytokine, which intricately controls a plethora of physiological and pathological processes during development and carcinogenesis. TGFβ1exerts anti-proliferative effects and functions as a tumor suppressor during early stages of tumorigenesis, whereas at later stages it functions as a tumor promoter aiding in metastatic progression through an autocrine TGFβ1 loop. For that, targeting TGFβ1is a valid approach to control cancer, at least in vitro [20].
Here, in the present study we are aiming to assess the efficiency of using CRISPR/Cas9-based approach to control breast and lung cancers via knockout of both CDK4 and TGFβ1, and activation of p107 genes.
MATERIALS AND METHODS
Cell line maintenance
Lung cancer cell line (A549) and breast cancer cell line (MCF-7) were obtained from the Holding Company for Biological Products and Vaccines (VACSERA, Giza, Egypt). All cells were seeded at a density of 104 cells/cm2 in 12-well plate, on RPMI 1640 media (GIBCO/Invitrogen Life Technologies, Carlsbad, USA) supplemented with 10% FBS (Hyclone, Logan, UT, USA) and 1% antibiotic mixture. Cells were cultured under standard laboratory conditions; at 37 °C and 5% CO2. MCF-7 and A549 cells were limited to use within the first 10 passages from the original purchased flask to control genomic drift due to instability. RPMI-1640 was changed every 3 days and cells were passaged whenever it reached 65–80% confluence or confluent?
CRISPR/Cas9 transfection
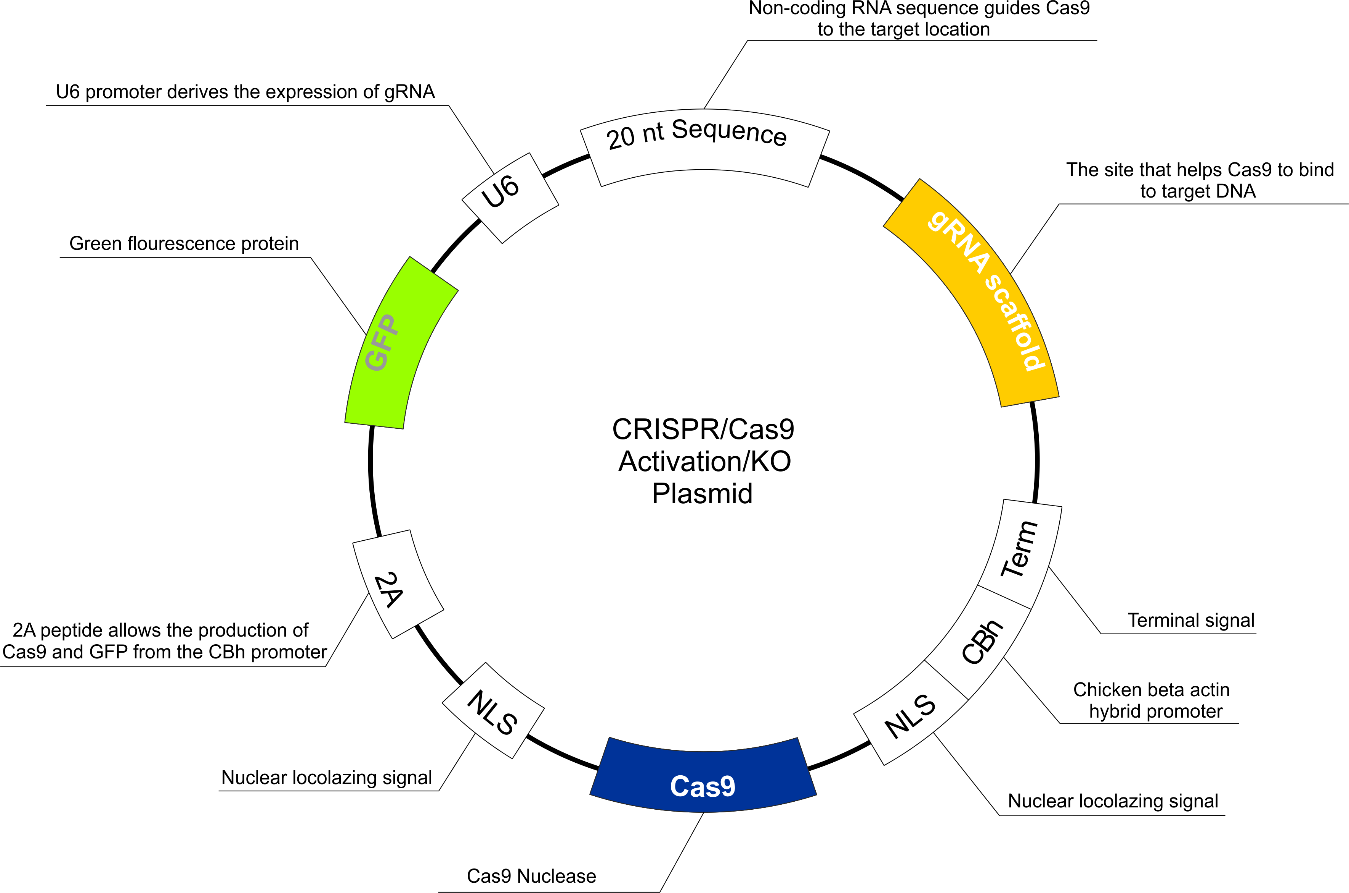
All CRISPR/Cas9 kits (p107 activation, CDK4 knockout, and TGFβ1 knockout) were purchased from Santa Cruz Biotechnology (USA) (Figure 1). The protocol of transfection was as follows: approximately 2 x 105 cells were seeded in a 6-well round U-bottom tissue culture plate on 3 mL of antibiotic-free standard growth medium per well 24 hours prior to transfection. Cells were grown to about 60-80% confluence, which was obtained normally after 24 hours. The following solutions were prepared: Solution A: For each transfection (and for each gene), 1μg (dissolved in 10 μL of deionized water) of plasmid solution was added to 140 μL of plasmid transfection medium to bring the final volume to 150 μL. The mix was pipetted up and down and incubated for five minutes at room temperature. Solution B: For each transfection (and for each gene), 10 μL of UltraCruz transfection reagent was diluted in 140 μL of plasmid transfection medium to bring the final volume to 150 μL. The mixture was pipetted up and down and incubated for 5 minutes at room temperature. The Plasmid DNA solution (Solution A) was added dropwise directly to the dilute UltraCruz transfection reagent (Solution B), vortexed immediately and incubated for 20 minutes at room temperature.
Prior to transfection, the old antibiotic-free medium was replaced with fresh media and the 300 μL of Plasmid DNA/UltraCruz transfection reagent mix (Solution A + Solution B) was added dropwise to the each well, and then gently mixed by swirling the plate. After that, the cells were incubated for 48 hours under normal laboratory conditions. Finally, the cells were harvested and resuspended in a 1 mL of fresh serum-containing medium to inactivate trypsin and cultured normally or stored at -20 °C for further analysis. The same cell lines were kept as control, where it was treated with transfection media and transfection reagents only.
Trypan Blue exclusion viable cell assay
After treatments, cells were trypsinized and resuspended in equal volumes of medium and Trypan blue (0.05% solution) and counted using a haemocytometer. Trypan blue dye (Invitrogen) exclusion was used to assess cell viability. Blue cells were considered non-viable, and unstained cells were considered viable.
RNA extraction
Total RNA was extracted from CRISPRized and unCRISPRized cells with the RNeasy kit (Qiagen, Hilden, Germany). RNA was treated with DNase I (Boehringer-Mannheim, Mannheim, Germany) for 50 min and purified according to the kit’s protocol. The quality and integrity of RNA were checked by spectrophotometry and ethidium bromide agarose gel electrophoresis.
First-strand cDNA synthesis
In a fresh tube, 200 ng of RNA and 1 μL of 25 μM random hexamer primer are mixed together and completed with RNase-free water to a final volume of 5 μl. The mixture was incubated at 72 °C for 3 min, and immediately placed on ice. Then, 2 μL of 5× SMARTScribe buffer, 1 μL of 10 mM deoxynucleotide triphosphate (dNTP) mix, 1 μL of 20 mM DTT and 1 μL of SMARTScribe Reverse Transcriptase (100 U/µl) (Clontech) were added to the mixture and mixed gently by up and down pipetting. The first-strand cDNA reaction mixture was incubated at 42 °C for 60 min and the reaction was terminated by heating at 70 °C for 15 min in a thermal block.
Table 1: The primers sequences used in the present study.
|
Primer name |
Sequence |
|
CDK4-F |
5′-GCGCCAGTTTCTAAGAGGCCTAGAT-3′ |
|
CDK4-R |
5′-CGGGTGTAAGTGCCATCTGGTAGCT-3′ |
|
p107-F |
5′-CAATGCTATAATGTGCCCAA-3′ |
|
p107-R |
5′-TAGGATTCCGCATACAAGAT-3′ |
|
TGFβ1-F |
5′-CATCCATGACATGAACCGACCCTT-3′ |
|
TGFβ1-R |
5′-ACGAAGTTGGCATGGTAGCCCTT-3′ |
Semi-quantification of gene expression
PCR was performed on all treated and untreated cells against the primers under study (Table 1). The following set-up was used and measured in triplicate: PCR mix 12.5 μL Master mix (Invitrogen), 0.5 μL forward primer (10 μM), 0.5 μL reverse primer (10 μM), and 1 μL cDNA (~ 30 ng) and dH2O up to 25 μL. The thermal profile set-up was: initial denaturation at 95 °C for 10 min; 35 cycles of 45 sec at 95 °C, 30 sec at (58 °C for CDK4, 57 °C for p107, and 60 °C for TGFβ1), and 75 sec at 72 °C; and then 5 min final extension at 72 °C. The PCR products were electrophoresed for 30 min on 1.6% agarose gel after being stained with ethidium bromide. The PCR assays were performed on the Uno II, Biometra. The relative expression of a specific mRNA compared to a control was calculated by measuring the bands intensity using Gel Pro analyzer, UK.
Protein extraction
Total protein extraction was performed using phenol extraction method; briefly, cells were centrifuged at high speed and resuspended in 3 mL of extraction buffer and stored at 4 °C. Just prior to use, 2-ME and 1 mM PMSF were added to pre-chilled buffer. The suspension was incubated for 10 min with continuous shaking at 4 °C. An equal volume of Tris-buffered phenol was then added, and the solution was again incubated on a shaker for 10 min at 4 °C. The aqueous and organic phases were separated by centrifugation at 13,000 rpm for 15 min at 4 °C. The phenolic phase was carefully recovered and re-extracted with equal volume of extraction buffer. The samples were vigorously vortexed and centrifuged for phase separation at 13,000 rpm for 15 min at 4 °C. The phenolic layer was transferred to a fresh tube for precipitation of proteins by addition of ammonium acetate (0.1 M) in cold methanol with subsequent overnight incubation at -20 °C. The precipitate/protein pellet was washed thrice with precipitation solution (stored at -20 °C), and final washing was performed with chilled acetone. The pellet was air-dried and then dissolved in rehydration buffer (7 M urea, 2 M thiourea and 2% (w/v) CHAPS).
Two-dimensional gel electrophoresis
The cellular proteins were solubilized in urea lysis buffer (7 M urea, 2% wt/vol CHAPS, and 50 mM DTT). The protein concentration was estimated by reagent compatible and detergent compatible (RC-DC) protein assay kit (Bio-Rad), and the readings were taken at 750 nm. To remove the contamination from the solubilized protein samples, urea/thiourea lysis buffer with ice-cold acetone was used. The precipitated proteins were suspended in rehydration buffer mentioned above.
Statistical analysis
Statistical significance was assayed by Student’s t test. The mean and standard error of the mean of the results of each experiment are shown in the figures. An asterisk indicates that p ≤ 0.05.
RESULTS
Knockout/activation by CRISPR/Cas9
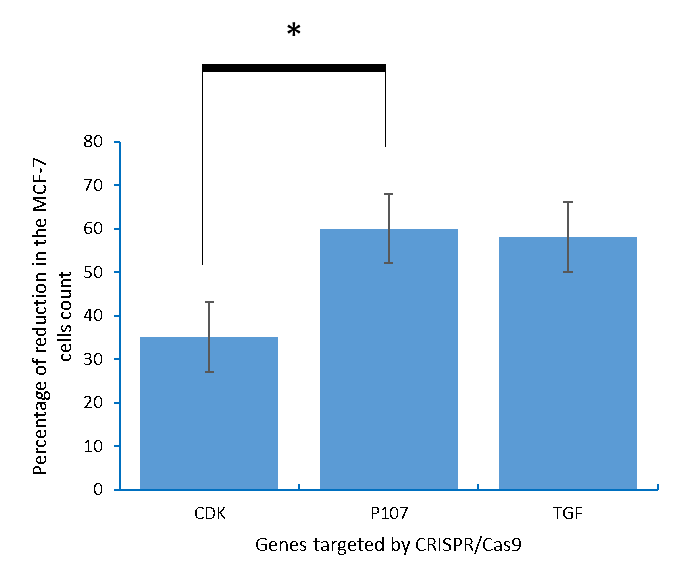
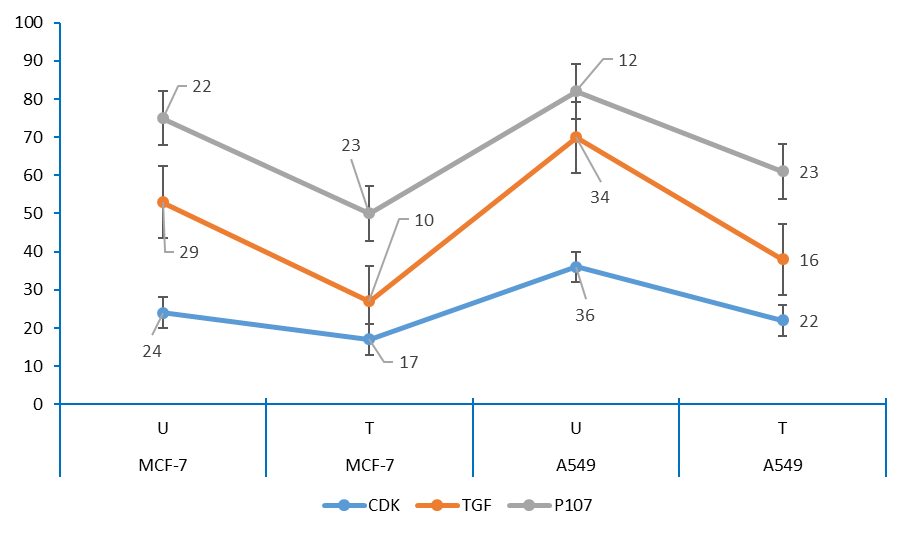
The present investigation aimed to control the proliferation of cancer cells that were resistant to traditional therapies by activation of p107 and knocking out of both CDK4 and TGFβ1. In the present study, two malignant cell lines (lung and breast) were used to study the impact of activation/knocking out of the target genes. Cell count was measured by trypan blue assay. Results obtained indicated that cell count was significantly reduced due to transfecting the two cell lines (p= .003). Breast cancer cells (MCF-7) were challenged with three plasmid transfections aiming to activate p107 and knockout both CDK4 and TGFβ1 (Figure 1-3). Transfecting MCF-7 cells with p107, CDK4, and TGFβ1 resulted in 63%, 35%, and 58.3% reduction in the cell count compared to non-treated ones, respectively.
Figure 1: The CRISPR/Cas9 activation/KO plasmid structure.
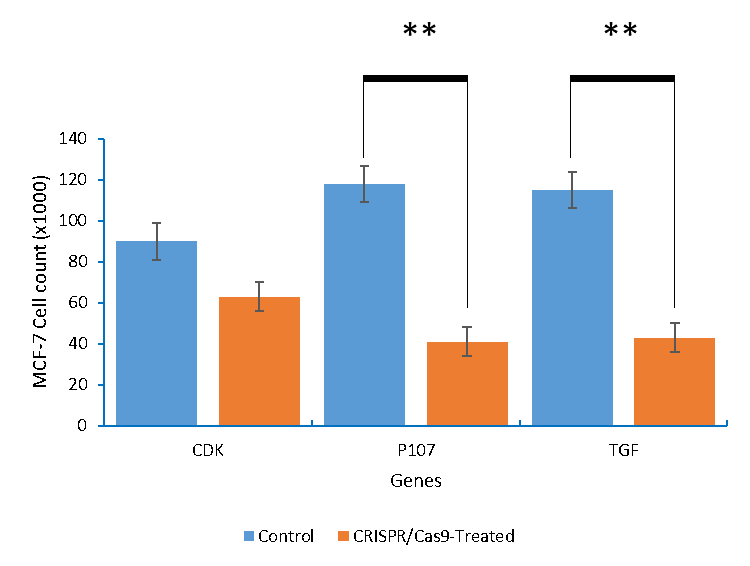
Figure 2: The MCF-7 breast cancer cells count after being knocking out of with CDK4 and TGFβ1, and activation of p107.
Figure 3: The percentages of the reduction in the MCF-7 breast cancer cells count caused by editing CDK4, p107, and TGFβ1 using CRISPR/Cas9.
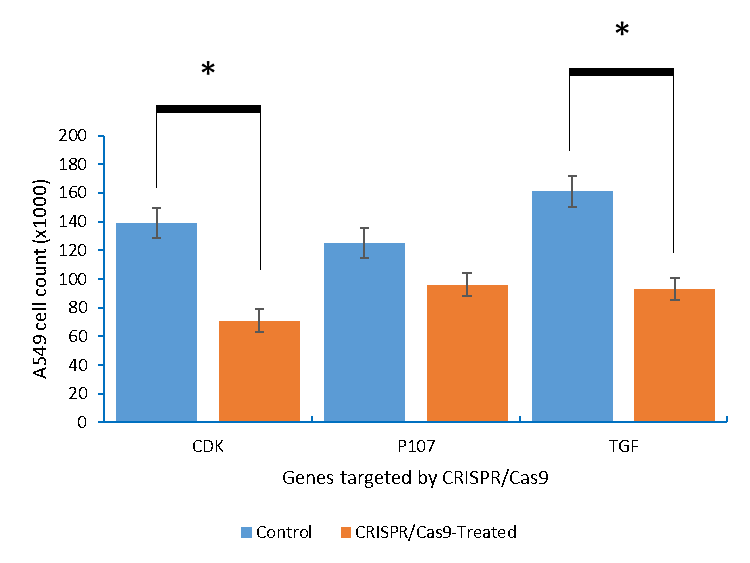
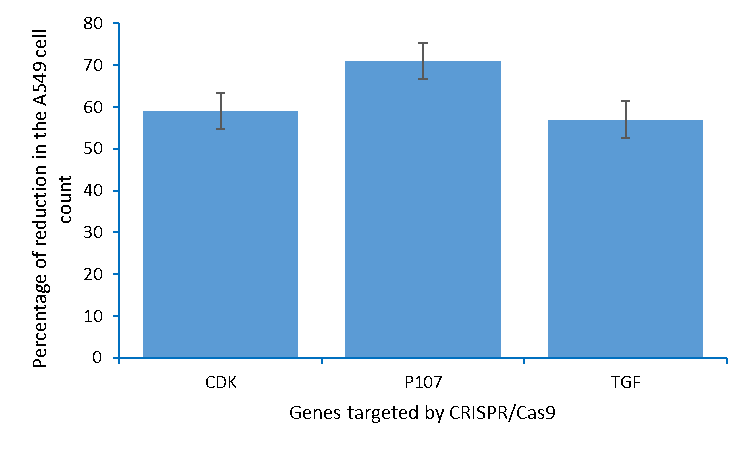
On the other hand, transfecting A549 lung cancer cells with p107, CDK4 and TGFβ1 resulted in 26.9, 41.6, and 45.1% reduction in the cell count, respectively (Figure 4 and 5).
Figure 4: The A549 lung cancer cells count after being knocking out of with CDK4 and TGFβ1, and activation of p107.
Figure 5: The percentages of the reduction in the A549 lung cancer cells count caused by editing CDK4, p107, and TGFβ1 using CRISPR/Cas9.
Detection of the edited genes
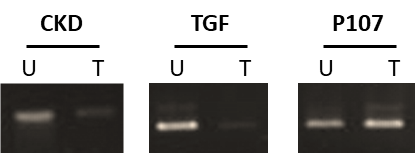
In the present study, MCF-7 and A549 cells were CRISPRized to edit CDK4, p107, and TGFβ1 in order to assess their role in the progression of these types of cancer. After activation of p107 and knocking out of both TGFβ1and CDK4, the presence/absence of these genes was detected using semi-quantitative PCR. Results obtained indicated that for breast cancer cells (Figure 6 and 8), knocking out both CDK4 and TGFβ1 affected the overall cell viability. Meanwhile, the activation of p107 caused the cells to commit suicide.
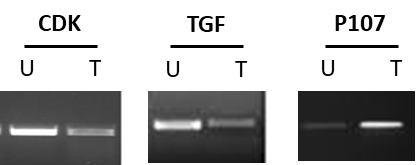
Lung cancer cells exhibited the same profile after being challenged with CRISPR/Cas9 to knockout TGFβ1and CDK4 and to activate p107 (Figure 7 and 8).
Our results showed that CRISPRising lung cancer cells to knockout CDK4 was not efficient in completely removing this gene. Although, a significant decrease (P= 0.0035) in the cells count was observed. Activation of p107 also resulted in decreasing the cell count compared to control.
Figure 6: Detection of CDK4, p107, and TGFβ1 in treated and untreated MCF-7 breast cancer cells by semi-quantitative PCR. T: treated with CRISPR/Cas9 and U: untreated.
Figure 7: Detection of CDK4, p107, and TGFβ1 in treated and untreated A549 lung cancer cells by qualitative PCR. T: treated with CRISPR/Cas9 and U: untreated.
Figure 8: The amount of DNA in control and each treatment MCF-7 and A549 cells.
2-D PAGE
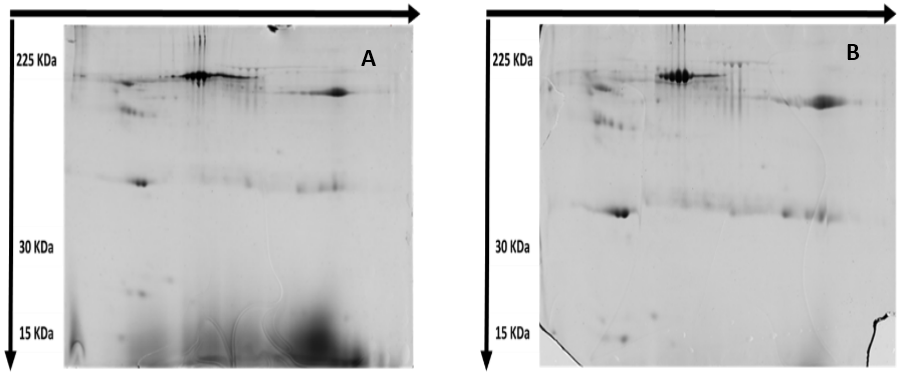
The two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) technique is a powerful proteomics technique aiming at separation of the complex protein mixtures; it can be used to discover proteins that are differentially expressed, and thus obtaining a set of potential biomarkers. The map was analyzed by the image created by Master 2D Elite Software. Two-dimensional protein electrophoresis experiments revealed some differences on the level of expressed proteins (Figure 9). There were some missing protein spots and, in the meantime, there were some de novo expressed protein spots.
TGF was also knocked out from MCF-7 and A549 cells via CRISPR/Cas9. Treated cells exhibited small amounts of the resemble protein. TGF-β1 is known for its multifaceted roles in breast cancer progression.
Figure 9: Protein expression map of (A) Lung cancer cell line (A549) and (B) Breast cancer cell line (MCF-7). One hundred-microgram proteins were separated by 2-DE using 13 cm pH 3-10 IPG strips and 12.5% homogeneous SDS-PAGE.
DISCUSSION
Knockout/activation of the target genes
P107 interacts with Retinoblastoma protein (RB) whose functions include DNA repair, telomere maintenance, chromosome condensation and cohesion, and silencing of repetitive regions [21], and thus, by editing (activating) p107, E2F-dependent transcription is suppressed, which might lead to cell death [21, 22]. This might explain the high percentage of reduction in the MCF-7 cell count reported in this study. Meanwhile, CDK family (such as CDK4) is found to be crucial in initiation and progression of breast cancer [23, 24] and in inducing apoptotic cell death [25]. Meanwhile, TGFβ1 knockout in the MCF-7 breast cancer cells is also resulted in nearly 60% reduction in the cell count. Although, TGFβ1 exerts tumor-suppressive effects that cancer cells must elude for malignant evolution, it also modulates processes such as cell invasion, immune regulation, and microenvironment modification that cancer cells may exploit to their advantage [26]. TGFβ1 switches its role to a tumor-promoting pathway by inducing endothelial to mesenchymal transition [27, 28]. However, the mechanism underlying such spatiotemporal regulation of TGFβ1 signaling needs in-depth investigation [29].
Activation of p107 might suppress tumorigenesis [30], and this gene plays a fundamental role in growth control [31]. Several researches indicated that p107 cooperates with the RB to suppress lung cancer [32, 33].
Knocking out of CDK4 resulted in 41.6% reduction in the cells count as preclinical studies have shown that CDK4 inhibition can produce rapid tumor regression and decrease in tumor burden in multiple human tumor xenograft models at high doses [34, 35]. It is also well known that overexpression of CDK4 is a potential unfavorable prognostic factor and mediates cell cycle progression by regulating the expression of p21 in lung cancer [32, 35]. Knocking out TGFβ1 also was critical for inhibiting the lung cancer cells growth, as it is a ubiquitous and essential regulator of cellular and physiologic processes including proliferation and differentiation [36]. It has a bi-functional tumor suppressor/oncogene role, and this might explain the 45.1% reduction in the lung cells count obtained in the present study. It has a bi-functional tumor suppressor/oncogene role [37], and this might explain the 45.1% reduction in the lung cells count obtained in the present study.
Amplification of the edited genes
Knocking out both CDK4 affected the overall cell viability as it suppresses the progression to S phase when downregulated [38]. It is amplified in approximately 20% of breast cancer cases, and the protein is over-expressed in 50% of cases [39]. TGFβ1 knocking out decreased the cells count in both cell lines as aberrant TGFβ1 signaling can lead to loss of growth inhibition [40]. Therapeutic targeting of the pro-oncogenic TGFβ1 responses is currently being explored as a potential therapy against certain invasive and metastatic cancer types [35]. Meanwhile, the activation of p107 caused the cells to commit suicide as it is involved in the negative regulation of cell cycle by interacting with E2F family members although in different combinations with respect to RB1[31].
TGFβ1 is overexpressed in various tumor tissues, promotes migration and invasion of cancer cells and known to induce EMT in a number of cancer cell types and promote lung adenocarcinoma migration and invasion [41]. Several studies have been conducted to study the effect of down regulation of TGFβ1 on lung cancer progression. Most of them used chemical substances such as sanguiin [42] and resveratrol [43]. Inhibition of CDK4 activity has turned out to be the most productive strategy for the discovery and design of novel anticancer agents specifically targeting the cell cycle.
Since CDK4 inhibitors were demonstrated to target proliferating cells, the induction of cell death, the partial knock-out of it have been proven as an effective approach in controlling the disease [38]. Meanwhile, a statistically significant inverse relationship between the histological grading (degree of malignant potential) and the expression of p107 in squamous cell carcinomas has been indicated, meaning that an increase in grading resulted in a significant decrease in protein expression, and this protein might compensate for chronic RB loss [44].
Specific protein detection
For p107 gene, CRISPR/Cas9-based activation has been performed, as this protein is important mediators of various cell processes including cell cycle progression, apoptosis and differentiation [32]. The downregulation of this gene has been widely correlated with different types of human cancers [31], and in regulating cell cycle. Meanwhile, CDK4, as an important regulator of the cell cycle, was knocked out from the malignant cells under study. Several researches indicated that misregulation of CDK4 activity can lead to cancer, as it regulates the cell cycle progression by regulating the G1-S checkpoint [34]. For that, eliminating the expression of this protein induce malignant cells to commit apoptosis by dysregulating key cellular pathways [35].
TGFβ1 is known for its multifaceted roles in breast cancer progression [45]. Treated cells exhibited small amounts of the resemble protein, although, the effect of TGFβ1 on cancer progression is variable: at early stages, TGFβ1 inhibits cancer progression, while at later stages it stimulates migration, invasion and metastasis [46]. However, the molecular basis of this “TGFβ1 paradox” is not entirely understood [47].
Conclusion
In the present study, the genomes of two cancer cell lines (MCF-7 and A549) were edited using CRISPR/Cas9 approach. TGFβ1 and CDK4 genes were knocked out while; p107 was activated to study their role in the process of cancer progression. Data obtained indicated a significant reduction in the cell count of both cell lines after being challenged with CRISPR cassettes. Meanwhile, the semi-quantitative PCR data revealed a partial downregulation of both CDK4 and TGFβ1, and upregulation of p107. Data obtained from 2D-PAGE indicated also the presence of de novo protein for p107 treatment while TGFβ1 and CKD4 proteins were absent. Generally, these data might indicate the efficacy of CRISPR/Cas9 in editing the genome of the specified cells.
Acknowledgment
The authors thank Mr. Basel El-Azab, College of Biotechnology, MUST, for his kind support during the conduction of this work.
Conflict of Interest
The authors declare that they have no competing interests.
Funding
This project has not received money from any funding bodies.
Authors’ contributions
HS*** designed the experiments and wrote the manuscript. DA**, NJ*, ST*, MF*, BM*, and AP* performed the experiments. SEA*, OAMS*, and AS* analyzed the data, EC** draw the illustrations, HT* revised the manuscript, AFA* revised charts, and ME*** critically revised the final manuscript. All authors read and approved the final manuscript.
*Participated equally
**Participated equally
***Participated equally
Ethics approval and consent to participate
Not applicable.
REFERENCES
- Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F (2017) Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 71(1): 96-108.
- Gao X, Reid MA, Kong M, Locasale JW (2017) Metabolic interactions with cancer epigenetics Mol Aspects Med 54: 50-57.
- Duruisseaux M, Esteller M (2018) Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol 51: 116-128.
- Hammarlund M, Hobert O, Miller DM 3rd, Sestan N (2018) The CeNGEN Project: The Complete Gene Expression Map of an Entire Nervous System. Neuron 99(3): 430-433.
- Heers H, Stanislaw J, Harrelson J, Lee MW (2018) Valproic acid as an adjunctive therapeutic agent for the treatment of breast cancer. Eur J Pharmacol 835: 61-74.
- Daniels B, Kiely BE, Lord SJ, Houssami N, Lu CY, Ward RL et al. (2018) Trastuzumab for metastatic breast cancer: Real world outcomes from an Australian whole-of-population cohort (2001–2016). Breast 38: 7-13.
- Seo JS, Kim A, Shin JY, Kim YT (2018) Comprehensive analysis of the tumor immune micro-environment in non-small cell lung cancer for efficacy of checkpoint inhibitor. Sci Rep 8(1): 14576.
- Wang S, Chen A, Yang L, Cai L, Xie Y, Fujimoto J et al. (2018) Comprehensive analysis of lung cancer pathology images to discover tumor shape and boundary features that predict survival outcome. Sci Rep 8(1): 10393.
- Osuoha CA, Callahan KE, Ponce CP, Pinheiro PS (2018) Disparities in lung cancer survival and receipt of surgical treatment. Lung Cancer 122: 54-59.
- Lian J, HamediRad M, Zhao H (2018) Advancing Metabolic Engineering of Saccharomyces cerevisiae Using the CRISPR/Cas System. Biotechnol J 13(9): e1700601.
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V et al. (2017) Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 358(6360): 234-238.
- O'Donnell KA (2018) Advances in functional genetic screening with transposons and CRISPR/Cas9 to illuminate cancer biology. Curr Opin Genet Dev 49: 85-94.
- Lee H, Zhou Y, Taylor DW, Sashital DG (2018) Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Mol Cell. 70(1): 48-59.e5.
- Kolli N, Lu M, Maiti P, Rossignol J, Dunbar GL (2018) Application of the gene editing tool, CRISPR-Cas9, for treating neurodegenerative diseases. Neurochem Int 112: 187-196.
- Kennedy EM, Cullen BR (2015) Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment. Virology 479-480: 213-220.
- Liu X, Zhang Y, Wang S, Liu G, Ruan L (2018) Loss of miR-143 and miR-145 in condyloma acuminatum promotes cellular proliferation and inhibits apoptosis by targeting NRAS. R Soc Open Sci 5(8): 172376.
- Gubern A, Joaquin M, Marques M, Maseres P, Garcia-Garcia J, Amat R et al. (2016) The N-Terminal Phosphorylation of RB by p38 Bypasses Its Inactivation by CDKs and Prevents Proliferation in Cancer Cells. Mol Cell 64(1): 25-36.
- Wang C, Jin H, Gao D, Wang L, Evers B, Xue Z et al. (2018) A CRISPR screen identifies CDK7 as a therapeutic target in hepatocellular carcinoma. Cell Res 28(6): 690-692.
- Witkiewicz AK, Chung S, Brough R, Vail P, Franco J et al. (2018) Targeting the Vulnerability of RB Tumor Suppressor Loss in Triple-Negative Breast Cancer. Cell Rep 22(5): 1185-1199.
- Abraham CG, Ludwig MP, Andrysik Z, Pandey A, Joshi M, Galbraith MD et al. (2018) ΔNp63α Suppresses TGFB2 Expression and RHOA Activity to Drive Cell Proliferation in Squamous Cell Carcinomas. Cell Rep 24(12): 3224-3236.
- Conklin JF, Baker J, Sage J (2012) The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun 3: 1244.
- Eyvani H, Moghaddaskho F, Kabuli M, Zekri A, Momeny M, Tavakkoly-Bazzaz J et al. (2016) Arsenic trioxide induces cell cycle arrest and alters DNA methylation patterns of cell cycle regulatory genes in colorectal cancer cells. Life Sci 167: 67-77.
- Jimenez RE, Hieken TJ, Peters MS, Visscher DW (2018) 12 - Paget Disease of the Breast, The Breast. [Fifth Edition], Elsevier, Amsterdam, Netherlands, 169-176.e3.
- Vogel VG (2018) 15 - Epidemiology of Breast Cancer, The Breast. [Fifth Edition], Elsevier, Amsterdam, Netherlands, 207-218.e4.
- Revathidevi S, Manikandan M, Rao AK, Vinothkumar V, Arunkumar G, Rajkumar KS, et al. (2016) Analysis of APOBEC3A/3B germline deletion polymorphism in breast, cervical and oral cancers from South India and its impact on miRNA regulation. Tumour Biol 37(9): 11983-11990.
- Plou J, Juste-Lanas Y, Olivares V, Del Amo C, Borau C, García-Aznar JM (2018) From individual to collective 3D cancer dissemination: roles of collagen concentration and TGF-β. Sci Rep 8(1): 12723.
- Chae YK, Chang S, Ko T, Anker J, Agte S, Iams W et al. (2018) Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci Rep 8(1): 2918.
- Huang Y, Zhang Z, Huang Y, Mao Z, Yang X, Nakamura Y, et al. (2018) Induction of inactive TGF-β1 monomer formation by hydrogen sulfide contributes to its suppressive effects on Ang II- and TGF-β1-induced EMT in renal tubular epithelial cells. Biochem Biophys Res Commun 501(2): 534-540.
- Xu Y, Niu J, Xi G, Niu X, Wang Y, Guo M, et al. (2018) TGF-β1 resulting in differential microRNA expression in bovine granulosa cells. Gene 663: 88-100.
- Zhang J, Xu K, Liu P, Geng Y, Wang B, et al. (2016) Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol Cell 62(6): 929-942.
- Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H (2004) Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev 18(23): 2952-2962.
- Luo Q, Li J, Cenkci B, Kretzner L (2004) Autorepression of c-myc requires both initiator and E2F-binding site elements and cooperation with the p107 gene product. Oncogene 23(5): 1088-1097.
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, et al. (2000) Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev 14(23): 3037-3050.
- Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. (2017) CDK4/6 inhibition triggers anti-tumour immunity. Nature 548(7668): 471-475.
- Dickson MA (2014) Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res 20(13): 3379-3383.
- Huang L, Jia J, Liu R (2013) Decreased serum levels of the angiogenic factors VEGF and TGF-β1 in Alzheimer's disease and amnestic mild cognitive impairment. Neurosci Lett. 550: 60-63.
- Dondeti MF, Talaat RM, El-Shenawy SZ, Khamiss OA (2017) Transforming growth factor (TGF-β1) gene polymorphisms in Egyptian patients with hepatitis B virus infection. Meta Gene 13: 5-12.
- Bertero T, Gastaldi C, Bourget-Ponzio I, Mari B, Meneguzzi G, Barbry P et al. (2013) CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell Death and Differ 20(6): 800-811.
- Liu J, Cheng Y, Wang X, Zhang L, Wang ZJ (2018) Cancer characteristic gene selection via sample learning based on deep sparse filtering. Sci Rep 8(1): 8270.
- Hoeman C, Shen C, Becher OJ (2018) CDK4/6 and PDGFRA signaling as therapeutic targets in diffuse intrinsic pontine glioma. Front Oncol 8: 191.
- Yang J, Zhang N, Gao R, Zhu Y, Zhang Z, Xu X, et al. (2018) TGF-β1 induced fascin1 expression facilitates the migration and invasion of kidney carcinoma cells through ERK and JNK signaling pathways. Biochem Biophys Res Commun 501(4): 913-919.
- Ko H, Jeon H, Lee D, Choi HK, Kang KS, Choi KC (2015) Sanguiin H6 suppresses TGF-β induction of the epithelial–mesenchymal transition and inhibits migration and invasion in A549 lung cancer. Bioorg Med Chem Lett 25(23): 5508-5513.
- Wang H, Zhang H, Tang L, Chen H, Wu C, Zhao M, et al. (2013) Resveratrol inhibits TGF-beta 1-induced epithelial-to-mesenchymal transition and suppresses lung cancer invasion and metastasis. Toxicology 303(1): 139-146.
- Madan E, Gogna R, Kuppusamy P, Bhatt M, Pati U, Mahdi AA (2012) TIGAR induces p53-mediated cell-cycle arrest by regulation of RB-E2F1 complex. Br J Cancer 107(3): 516-526.
- Zhao Y, Wang X, Wang Q, Deng Y, Li K, Zhang M, et al. (2018) USP2a Supports Metastasis by Tuning TGF-β Signaling. Cell Rep 22(9): 2442-2454.
- Shirvani-Farsani Z, Behmanesh M, Mohammadi SM, Naser Moghadasi A (2015) Vitamin D levels in multiple sclerosis patients: Association with TGF-β2, TGF-βRI, and TGF-βRII expression. Life Sci 134: 63-67.
- Ros XR, Vermeulen L (2018) Turning Cold Tumors Hot by Blocking TGF-β. Trends Cancer 4(5): 335-337.
Copyright: Sabit H, et al. ©2019. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Sabit H (2019). CRISPR/Cas9-based Editing of CDK4, p107, and TGFβ1 in Human Breast and Lung Cancer Cells. Oncogen 2(3): 15.
 Abstract
Abstract  PDF
PDF